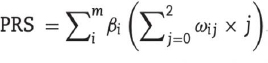All published articles of this journal are available on ScienceDirect.
The Study of the COVID-19 Outbreak: A Review
Abstract
COVID-19 was declared a pandemic in 2020 and spanned a three-year period, causing devastating effects across the globe. The death toll from the infection reached millions, with medical experts and government officials worldwide working tirelessly to control its spread. Symptoms from the virus ranged from mild (i.e., fever and cough) to severe (i.e., respiratory failure and multi-organ dysfunction), creating difficulties in tracking its progression and developing appropriate treatments. The aim of this article is to provide a comprehensive review of the SARS-Cov-2 virus, the cause of COVID-19, and its varied characteristics studied throughout the pandemic, including its structure, common symptoms, and the numerous treatment options made available. Viral and host genetics are described as well, as multiple studies have linked molecular variants to differing degrees of disease severity. Polygenic risk scoring (PRS) has been an approach used for the determination of risk for severe outcomes, assisting with the identification of significant genetic variants and high-risk population groups.
1. INTRODUCTION
There are multiple types of coronaviruses, each infecting different animals and involving numerous modes of host cell entry. They can be divided into three genera (I, II, and III) based on recent genome sequence analysis [1]. Group I includes animal pathogens, as well as the human coronaviruses HCoV-229E and NL63, which are responsible for various respiratory infections. Group II also includes animal pathogens, which are of veterinary relevance, and human coronaviruses OC43 and HKU1, also causing respiratory infections. Group III, as of now, only includes avian coronaviruses [2]. There is currently a debate regarding whether SARS-CoV defines a new group of coronaviruses or whether it is a distant member of Group II (Table 1) [3, 4].
2. THE VIRAL STRUCTURE, MECHANISM OF ENTRY, LABORATORY DIAGNOSTICS, AND DISEASE STAGES
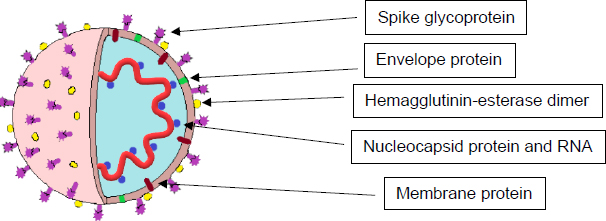
SARS-CoV-2, the cause of COVID-19, is a positive-sense, single-stranded RNA virus surrounded by an envelope made of glycoprotein. Under an electron microscope, it has a crown or “corona”-like morphology, which is what established the name “coronavirus” for this family [4]. The genome is comprised of roughly 30,000 nucleotides, which encode all its necessary proteins, including four structural proteins and numerous non-structural proteins (Fig. 1). The structural proteins include the following:
Table 1.
| Group | Virus |
| I | 229E (human coronavirus) |
| TGEV | |
| PRCoV | |
| Canine corornavirus | |
| FeCoV | |
| FIPV | |
| NL-63 (human coronavirus) | |
| II | OC34 (human coronavirus) |
| MHV | |
| Sialodacryoadenitis coronavirus | |
| Hemagglutinating encephalomyocarditis virus | |
| BCoV | |
| HKU1 | |
| SARS-CoV | |
| III | IBV |
| Turkey coronavirus |
2.1. Nucleocapsid (N) Protein
Within the protein shell capsid, there is the N-protein, which binds to the viral RNA via the N-terminal of the protein, forming the nucleocapsid. This process allows the virus to hijack human cells during infection and use their cellular machinery to create new viral particles. It is a vital protein involved with viral replication and transcription and ultimately completes the viral formation [5].
2.2. Spike (S) Glycoprotein
The integration of spike (S) glycoprotein over the surface of the virus allows it to mediate its attachment to host cell angiotensin-converting enzyme 2 (ACE2) receptors and fusion between the viral and host cell membranes, facilitating entry [6]. It is made of three identical chains of 1,273 amino acids each and two well-defined protein domain regions: S1 and S2 subunits. These are associated with cell recognition and fusion of the viral and cell membranes, respectively [7]. It has been observed in some coronavirus strains that the expression of the S protein can also mediate cell-to-cell fusion between infected and adjacent, uninfected cells. This forms giant multinucleated cells, or syncytia, which are potentially used as a strategy for the virus to spread directly between cells while avoiding countering antibodies [7, 8].
2.3. Envelope (E) Protein
This small membrane protein is made of roughly 76-109 amino acids and is a minor component of the virus particle. Nevertheless, it plays an important role in virus assembly, membrane permeability of the host cell membrane, and viral-host cell interactions [9]. The majority of this protein is located at the site of intracellular trafficking, such as the endoplasmic reticulum and the Golgi apparatus, where it assists with viral assembly, budding, and maturation [10].
2.4. Membrane (M) Protein
This is the most abundant protein found on the viral surface and determines the shape of the viral envelope. It is believed to be the central organizer for assembly via interactions with the other major proteins [11].
- Interaction between S and M proteins is necessary for retention of the S protein at the site of intracellular trafficking to ensure incorporation into new viral particles [12].
- Binding M with N proteins stabilizes the nucleocapsid as well as the internal core of virions, which ultimately allows the completion of the viral assembly [13].
- M and E proteins together make up the viral envelope, and their interaction allows for the adequate production and release of virus-like particles (VLPs) [14].
Another non-structural protein is the Hemagglutinin-esterase dimer (HE), located on the viral surface. It is not required for replication; however, it may be involved with viral entry into the host cells and appears to be important for infection [15].
The mechanism of viral entry and replication in the host cell involves numerous steps. The spike protein is first attached to the ACE2 receptors located on the surface of many human cells, allowing viral entry. The S protein is then cleaved by host proteases between the S1 and S2 subunits. In a later stage, the S2 domain is cleaved, releasing a fusion peptide that leads to the activation of membrane fusion. Human cells will generally ingest the virus via endocytosis. Once in the endosome, a three-step process has been hypothesized as the method for membrane fusion. This involves receptor-binding, induced conformational changes in the S glycoprotein, and cathepsin L proteolysis within the endosome via intracellular proteases [16]. This is followed by the opening of the endosome to allow the virus entry into the cytoplasm and uncoating of the viral nucleocapsid. A second hypothesis for viral entry involves a two-step method where the virus binds to a host cell surface receptor by its S1 subunit, resulting in the S glycoprotein being cleaved by the host proteases. This causes fusion of the viral and host cell membranes at low pH via the S2 subunit [17]. Once the virus has entered the cytoplasm, the genetic material can then be replicated via the replication/transcription complex (RTC). This complex is part of the viral genome and is made of non-structural proteins. Replicase proteins are translated from the genome, allowing for the generation of full-length negative sense RNA strands. These are then used as templates for new full-length genomes. The required structural proteins are generated, and the newly created virions are exported from the host cell by transportation to the cell membrane in vesicles and secreted via exocytosis. These new virions are then able to infect other host cells (Fig. 2) [18].
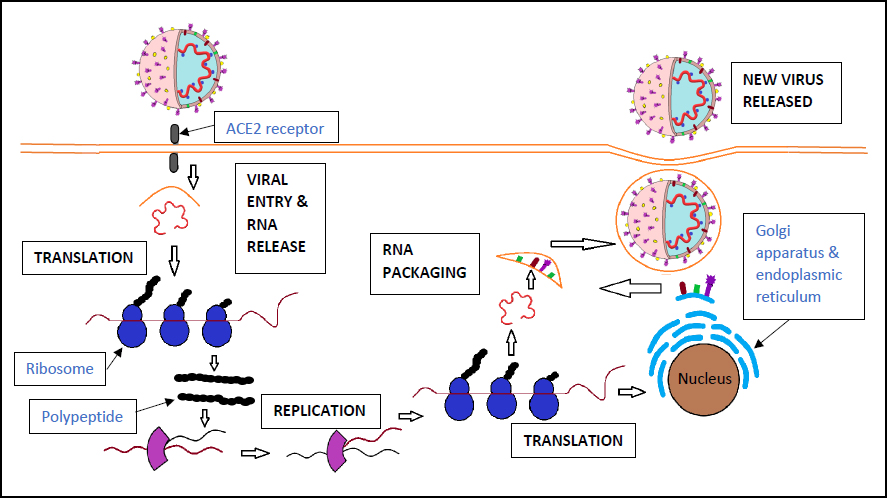
The mechanism of SARS-Cov-2 entry and replication. The virus enters the host cell via the ACE2 receptor and releases its RNA. The RNA is translated into polypeptides, which then undergo proteolysis to release active proteins. This allows for RNA replication and protein translation, creating new viral particles to be released from the host cell [16-18].
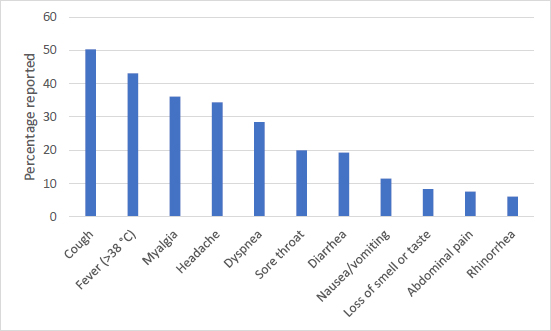
Most commonly associated symptoms with COVID-19 [19].
Other strains of coronavirus generally only cause mild infection, according to case reports from the Chinese Center for Disease Control and Prevention. COVID-19 symptoms, however, can range from mild to severe to critical. The most commonly experienced COVID-19 symptoms are cough, fever, and myalgia, according to the CDC of the United States (Fig. 3) [19]. In the first part of the pandemic, 81% of patients tended to have mild symptoms, exhibiting either no or mild pneumonia symptoms. Up to 14% of patients had severe disease, exhibiting dyspnea, hypoxia, or >50% lung involvement within the first 2 days of infection. The critical disease was found in 5% of patients who exhibited respiratory failure, shock, or multi-organ dysfunction [20]. Patients were hospitalized when they experienced severe or critical disease. However, it is estimated that around 33% of infected patients remained asymptomatic [21]. Overall fatality rates varied by location and across different risk groups, with an estimation to be between 0.1-6.0% case-fatality by country. It was initially thought to be much higher due to the larger number of asymptomatic cases that were not diagnosed [22, 23]. Fatality rates have progressively dropped due to the availability of new vaccines and treatments; however, severe cases still result in hospitalization with critical disease.
COVID-19 has multiple clinical phases that can be used to monitor its progression. While it was initially believed to be only a respiratory disease, it has become more evident that it is a multi-organ and heterogeneous illness. As the disease progresses and the severity increases, the disease stage can be determined using objective and molecular criteria. Knowing the disease data can assist in clinical decisions for patient management, improved prognosis, and appropriate treatment selections. Cordon-Cardo et al. (2020) described the characteristics of the stages, including the therapeutic interventions (Table 2). The first stage involves diagnostic tests, such as SARS-CoV-2 detection and viral load (RT-qPCR, qLAMP). The other three stages involve disease monitoring for inflammatory markers, cytokine panels, thrombosis markers, and antibody tests (quantitative, neutralization, etc.) [24].
| Stage | Characteristics | Treatments |
|---|---|---|
| Stage 1: Asymptomatic | Viral entry and replication | - |
| Stage 2: Mild/Moderate | Viral dissemination | Anti-virals, antithrombotics, steroids, immunomodulators, convalescent plasma, hyperimmune serum, and clonal antibodies |
| Stage 3: Severe | Multisystem inflammation | Respiratory support, antithrombotics, steroids, immunomodulators, convalescent plasma, hyperimmune serum, clonal antibodies, and mesenchymal stem cells |
| Stage 4: Critical | Endothelial damage and thrombosis | Respiratory support, antithrombotics, steroids, immunomodulators, and mesenchymal stem cells |
COVID-19 diagnosis is typically performed using reverse transcriptase-polymerase chain reaction (RT-PCR) assay, chest CT, or both, depending on access to local viral testing and the time of diagnosis in the course of the disease. Chest CT is most effective in disease monitoring for patients who have progressed to pneumonia. In patients with COVID-19 pneumonia, ground-glass opacities were observed on chest CT more frequently than in patients with non-COVID-19 pneumonia [25, 26]. RT-PCR is used to generate cDNA from SARS-CoV-2 RNA extracted from respiratory samples. Common SARS-CoV-2 gene targets include the envelope, nucleocapsid, spike, RNA-dependent RNA polymerase, and ORF1 genes [27]. Multiple different primer-probe sets for detection and test kits have been developed with variable sensitivity [28]. The viral RNA obtained from the samples is detected via PCR, where the RNA undergoes multiple replications until it can be detected with a fluorescent signal. The fewer cycles required for detection indicate a higher viral RNA load. When the number of cycles is less than 40, the test is considered positive. However, a positive PCR result indicates the presence of viral RNA and is not necessarily a viable virus [27].
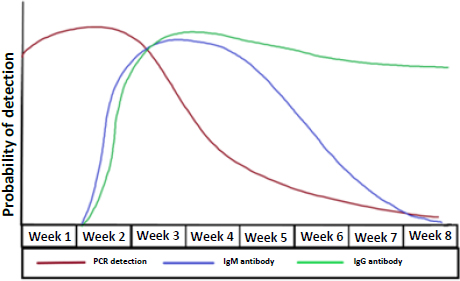
Generally, the viral RNA becomes detectable as early as day 1 of symptoms and peaks within the first week of onset. Positivity usually starts to decline by week 3 [29]. It also depends on the type of sample obtained, as a sputum sample may remain positive for longer than a nasopharyngeal swab. Highest positivity is found in bronchoalveolar lavage specimens (93%), followed by sputum (72%), nasal swab (63%), and pharyngeal swab (32%) [30]. An indirect method to detect a positive COVID-19 infection is to measure the immune response to the virus. Measuring serology can be useful in cases where the patients present beyond the first 2 weeks of onset due to a milder course. It can also help identify how widespread COVID-19 is within a community. IgM and IgG antibodies can be detected as early as the fourth day of symptom onset, and higher levels are observed in the second and third week of illness. Afterwards, IgM levels begin to decline and almost disappear by week 7, whereas IgG levels will continue to persist beyond 7 weeks (Fig. 4) [31, 32].
Studies have shown that progressive respiratory failure is the primary cause of death in those infected with the SARS-Cov-2 virus. This is due to the development of acute respiratory distress syndrome (ARDS)-like symptoms; however, COVID-19 pneumonia is a specific type of respiratory disease. ARDS is an acute, life-threatening inflammation of the lung due to infection, trauma, or other inflammatory conditions. This excessive inflammation leads to alveolar damage and increased permeability of endothelial and epithelial cells, causing decreased respiratory compliance. These cellular changes result in the accumulation of protein-rich fluid in the interstitium and air space, which causes impaired gas exchange and varying levels of hypoxemia. This impairment of the lung microvascular barrier is central to the pathogenesis of ARDS [33, 34]. Chemokines and cytokines have also been shown to contribute to lung injury, as they lead to the recruitment of various types of immune cells, such as neutrophils. These immune cells are important for resolving the cause of the inflammation; however, they can lead to significant damage to the normal lung cells as well [35].
ARDS in patients with COVID-19 have been observed to have different physical changes than in patients with ARDS from other causes. One study compared the lungs of seven patients who died from COVID-19 to the lungs of seven patients who died from ARDS secondary to an influenza infection and 10 uninfected control lungs. The lungs of those who died of COVID-19 or influenza-associated respiratory failure were found to have diffuse alveolar damage in the peripheral lung with perivascular T-cell infiltration. However, COVID-19 lungs also showed distinctive vascular features involving severe endothelial injury associated with an intracellular virus and disrupted cell membranes. There was also widespread thrombosis with microangiopathy. Alveolar capillary microthrombi was found to be 9x more prevalent in patients with COVID-19 than those with influenza. There was also a 2.7x increase in new vessel growth among COVID-19 patients, predominantly through intussusceptive angiogenesis [36].
Comorbidities are quite commonly seen in patients infected with COVID-19, with hypertension being the most common, followed by diabetes and coronary heart disease. Risk factors, such as older age, high lactate dehydrogenase levels, and a D-dimer level greater than 1 μg/mL, were also indicative of a poor prognosis in the early stage [37, 38]. Other studies also found independent risk factors that were associated with no improvement in these patients, including male sex, a severe COVID-19 condition, expectoration, muscle ache, and decreased albumin [39]. Additionally, other particular laboratory findings have been associated with worse outcomes, including lymphopenia, thrombocytopenia, and elevated liver enzymes (Table 3) [40, 41].
| Laboratory feature | Abnormal value | Normal range |
|---|---|---|
| D-dimer | >1000 ng/mL | <500 ng/mL |
| CRP | >100 mg/L | <8.0 mg/L |
| LDH | >245 units/L | 110-210 units/L |
| Troponin | >2x the upper limit of normal | Females: 0 -9 ng/L Males: 0-14 ng/L |
| Ferritin | >500 mcg/L | Females: 10-200 mcg/L Males: 30-300 mcg/L |
| CPK | >2x the upper limit of normal | 40-150 units/L |
| Absolute lymphocyte count | <800/μL | 1800-7000/μL |
| Platelet count | <23-31 x 109/L | 150-450 x 109/L |
| AST and ALT | >3x the upper limit of normal | AST: 10-40 units/L ALT: 7-56 units/L |
While initially believed to be exclusively a respiratory disease, COVID-19 has been observed to involve more than just the throat and the lungs. The virus has been observed to have a significant impact on the cardiovascular system, leading to various conditions including myocarditis, myocardial injury, scar formation, arrhythmias, heart block, and vascular occlusion due to local thrombus formation or embolism, which can lead to cardiac death [42-44]. Other potential systems include the brain and the kidneys, resulting in multi-organ failure. It has been hypothesized that, especially in the later complicated stages of the disease, COVID-19 can be considered an endothelial disease. The reasoning for this takes into consideration how the vascular endothelium cells not only direct the circulating blood to the tissues but also assist with maintaining homeostasis in the body. This includes regulating an array of functions, such as hemostasis, fibrinolysis, inflammation, oxidative stress, vascular permeability, and vasculature structure. These functions normally contribute to coordinating host defenses and maintaining overall homeostasis; however, they can also contribute to disease if defense mechanisms overreach during an illness and turn against the host. Such is the case of COVID-19, which triggers an increase in cytokines and protein proinflammatory mediators, triggering endothelial functions towards a defensive mode. Continuing down this path can lead to a cytokine storm with positive feedback loops maintaining the cytokine production that overpowers the counter-regulatory mechanisms, resulting in significant damage to the host [45].
SARS-Cov-2 has been shown to bind to the receptor ACE2 on cell surfaces to gain access to the host cells. This receptor plays a functional role in the renin-angiotensin-aldosterone system (RAAS), which is a signalling pathway involved with the homeostatic regulation of vascular functions. This includes control over systemic and local blood flow, blood pressure, natriuresis, and trophic responses to many different stimuli. Angiotensin II (Ang II) is the main effector molecule of the system and binds to the ACE2 receptor [46, 47]. SARS-Cov-2 competes with Ang II for this receptor. ACE2 expression is seen in numerous types of cells, including respiratory epithelial cells, myocardial cells, oesophagus epithelial cells, tongue epithelial cells, and enterocytes from the ileum, to name a few [48-50]. This variety in cell types expressing ACE2 can help explain why patients exhibit a vast array of symptoms when infected with SARS-Cov-2 due to the ability of the virus to diffuse throughout the body.
3. PAST/CURRENT TREATMENTS AND VACCINES
By March 2020, COVID-19 had become so widespread it was characterized as a global pandemic. As lockdowns increased to slow the spread, efforts increased to develop treatment options for infected patients. Initially, no specific treatments showed high efficacy for treating the infection. Beyond the use of interventional therapies to treat the respiratory issues associated with the infection, including high-flow nasal oxygen, prone positioning and fluid management [51], classes of drugs mainly used as potential treatments included antiviral agents, immunomodulators, inflammation inhibitors, low-molecular-weight heparins, plasma, and hyperimmune immunoglobulins. Examples of these treatments include the following:
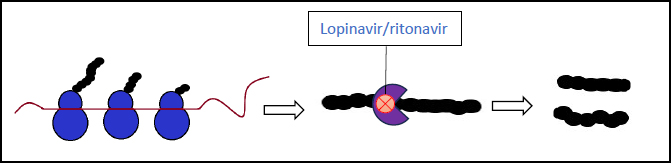
3.1. Lopinavir-Ritonavir (Kaletra)
These two combined antivirals are protease inhibitors that inhibit viral replication (Fig. 5). They were mainly used in COVID-19 patients in the early stages with mild symptoms, being managed at home or in the hospital. Previous experiences with SARS-CoV-1 and MERS infections have suggested that this drug may improve the outcomes in some patients. However, a randomized trial study for severe COVID-19 cases showed no significant clinical improvement with the use of lopinavir-ritonavir therapy [52, 53].
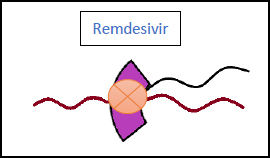
3.2. Remdesivir (Veklury)
This antiviral belongs to the class of nucleotide analogues and can be used in moderate and severe cases of COVID-19 (Fig. 6). Multiple studies have been conducted to examine its efficacy, which have concluded the following results:
- Grein et al. (2020) analyzed the use of remdesivir in a small cohort of severe COVID-19 patients. A total of 68% of patients who followed up after treatment were found to have clinical improvement [54].
- Goldman et al. (2020) obtained the first results of the phase 3 trial on the use of remdesivir in hospitalized patients, where patients with severe COVID-19 were administered the drug for either 5 or 10 days. By day 14 after initiation of treatment, clinical improvement was found in 64% of patients in the 5-day treatment group and 54% in the 10-day treatment group. While there was no great distinction between a 5-day and 10-day treatment course, it is suggested that patients undergoing mechanical ventilation could benefit from the 10-day treatment. However, further studies on high-risk groups are needed to determine the shortest period of therapy [55].
- Beigel et al. (2020) did a randomized controlled trial with 1,059 patients, with 538 assigned to remdesivir and 512 on placebos. Patients assigned remdesivir were shown to have an average hospitalization time of 11 days with a mortality estimate of 7.1%, compared to 15 days with an 11.9% mortality estimate among those on placebos [56].
These studies have suggested that the use of remdesivir can prevent the progression of a COVID-19 infection, as well as shorten the recovery times for hospitalized patients.
3.3. New Molecules
Two molecules were developed by Dai et al. (2020) that are capable of blocking molecules 11a and 11b, which are protease enzymes that allow replication of SARS-CoV-2. Both molecules showed anti-COVID-19 activity in cell culture, and neither were observed to cause significant cytotoxicity. Further investigation was conducted in animal experiments to determine extended pharmacological potential. Both showed good pharmacokinetic properties, such as high bioavailability and half-life, and further studies on molecule 11a showed no obvious toxicity when conducted on animals, suggesting it might be a good candidate for human clinical trials [57].
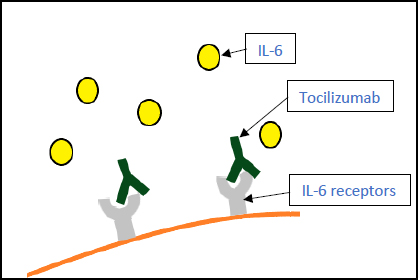
3.4. Tocilizumab (Actemra)
An antibody directed against the interleukin-6 (IL-6) receptor, tocilizumab, was previously the most used drug against COVID-19 (Fig. 7). Multiple studies were conducted to determine its efficacy, and the results were as follows:
- A study was conducted by the Agenzia Italiana de Farmaco (AIFA), 2020 to determine its safety and efficacy. Results suggested tocilizumab could significantly reduce mortality at one month but had less of an impact on early mortality (i.e., 14 days) [58].
- Guaraldi et al. (2020) conducted a retrospective observational cohort study that included adults with severe COVID-19 pneumonia who were hospitalized at a tertiary center. The study evaluated the efficacy of tocilizumab treatment when combined with usual care in reducing mortality and its impact on the likelihood of invasive mechanical ventilation when compared with those receiving standard treatment. It was concluded that administration of tocilizumab, both intravenous and subcutaneous, may help reduce the risk of invasive mechanical ventilation or death in patients with severe COVID-19 pneumonia [59].
- Another study published on the AIFA website (2020) was conducted to determine the effectiveness of early administration of tocilizumab in patients with COVID-19 pneumonia. There were no significant differences observed in the number of ICU admissions and 30-day mortality rate, and it was therefore concluded that early administration of the drug did not provide any clinical benefit for the patients [60].
3.5. Anakinra (Kineret)
Anakinra is an inhibitor of IL-α and IL-β proinflammatory cytokines (Fig. 8). It was suggested that treating the hyperinflammation of COVID-19 patients could improve mortality rates. King et al. (2020) reviewed many studies that used anakinra to target hyperinflammation in COVID-19. They found some success in treating the macrophage activation syndrome caused by inflammatory conditions overall. It was important to take the dose and administration of anakinra into consideration due to its short plasma half-life. Studies have shown that using the subcutaneous route, for example, could guarantee an adequate plasma concentration with a bioavailability ranging from 80 to 95% [61].
3.6. Baricitinib (Olumiant)
This is an inhibitor of Janus kinases (JAK), which are enzymes involved in the transduction of the cytokine-mediated signal (Fig. 9). Overstimulation of this pathway can potentially lead to hyperinflammation, as seen in many COVID-19 patients. Cantini et al. (2020) conducted an observational study in hospitals with moderate COVID-19 pneumonia patients to evaluate the effectiveness and safety of 2-week treatments with baricitinib plus antivirals in comparison with the standard of care therapy. It was observed that the 2-week case fatality rate was significantly lower in patients treated with baricitinib than the controls. This treatment may have also reduced the number of ICU admissions and deaths [62].
3.7. Corticosteroids
These are immunosuppressants that can be used in disease states where the body is hypersensitive and attacking the cells of the host (Fig. 10). A study was conducted in the United Kingdom called the randomised RECOVERY trial to determine which drugs were the most effective for treating adults hospitalised with COVID-19. The drugs in the study included low-dose dexamethasone (a corticosteroid), lopinavir-ritonavir, hydroxychloroquine (an immunomodulator), azithromycin (an antibiotic), and tocilizumab (a monoclonal antibody). When examining the control group, who only received standard treatments, it was observed that 28-day mortality was higher in those who needed ventilation and intermediate in those who only required oxygen. In the dexamethasone treatment group, mortality was observed as one-third lower in ventilated patients and one-fifth lower in those requiring only oxygen [63]. The World Health Organization (WHO) recommended that corticosteroids should not be used in treatment for non-severe COVID-19 cases as they provide no benefit [64].
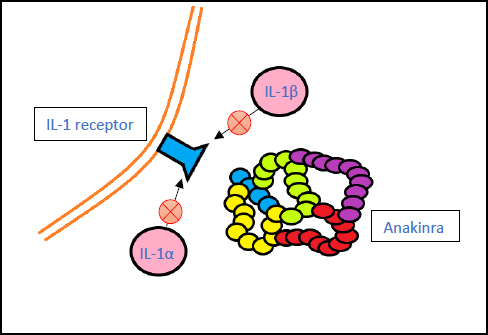
Anakinra is an immunosuppressive drug. It functions by inhibiting the actions of IL-α and IL-β proinflammatory cytokines on cellular receptors [61].
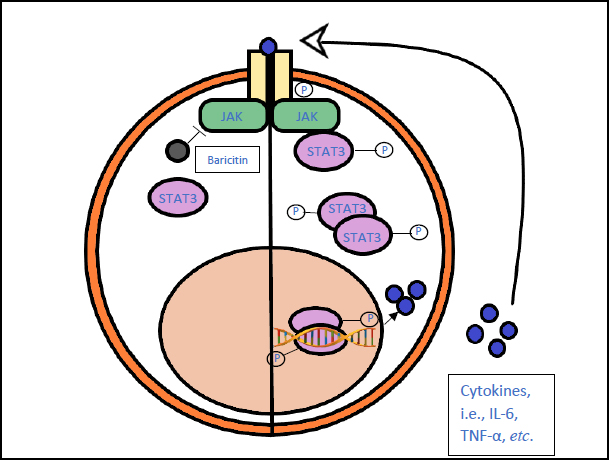
Baricitinib is a JAK inhibitor, preventing the increased production of cytokines that leads to hyperinflammation [62].
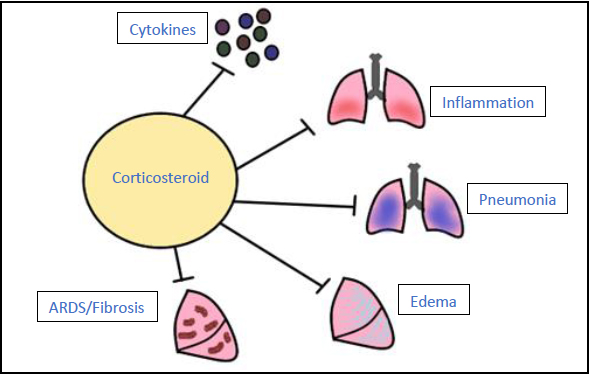
Corticosteroids are immunosuppressants that inhibit the body’s innate immune response. For a COVID-19 infection, corticosteroids can be given to prevent the increased production of cytokines and inflammation of the lungs, reducing the risk for pneumonia, fluid retention, and development of ARDS and fibrosis [63, 64].
3.8. Anticoagulants
These are medicines that help prevent blood clots. Progressive alteration of certain inflammatory and coagulative parameters can occur in the late stages of COVID-19 infections. These include increased degradation of fibrin, leading to increased levels of D-dimer, consumption of coagulation factors, and thrombocytopenia, to name a few. Therefore, at this stage of the disease when it becomes increasingly important to contain hyperinflammation due to the risk of blood clots, anticoagulant drugs such as non-fractionated low-molecular-weight-heparin (LMWH) or unfractionated heparin can be used to reduce this risk. A retrospective analysis was conducted in 2020 that examined many cases of severe COVID-19 pneumonia patients. For those who had observable activation of their coagulation cascade, if they were administered heparin for at least 7 days it was suggested to be an advantage in terms of survival. LMWH can also be used as a prophylaxis in the earlier stages of the disease when the patient suffers from hypomobility due to the illness to prevent the formation of venous thromboembolism [65].
3.9. Therapeutic Antibodies
These are SARS-COV-2 antibodies taken from blood samples of patients who had recovered from COVID-19 to serve as an alternative therapy for patients who were subsequently infected. It was estimated that the required dose of antibodies needed to treat infected patients required the removal of antibodies from at least three recovered individuals to reach the optimal concentration. There is limited data on the effectiveness of these hyperimmune immunoglobulins as a treatment option due to the short period of time between the beginning of the pandemic and when this treatment was under consideration. The following studies were conducted with limited results:
- One of the first studies in 2020 used plasma for treatment of infected patients. The results initially reported improvement in the clinical course and overall survival of the patients after additional administration of plasma and hyperimmune immunoglobulins. However, these findings need to be confirmed through randomized clinical trials (RCTs) [66].
- A trial evaluated the safety and efficacy of using donated plasma with SARS-CoV-2 antibodies as a treatment option for hospitalized patients. The study was terminated due to insufficient eligible participants; however, the results obtained did not show significant improvement with this treatment option [67].
Overall, the effectiveness of using the plasma of recovered COVID-19 patients to treat patients subsequently infected was not shown to be significant. This treatment, plus the previously described options, indicated the necessity of developing newer, more effective treatment options.
3.10. Molnupavir (Lagevrio)
This is a nucleoside analog that inhibits SARS-CoV-2 replication. Multiple studies have been performed on the effectiveness of this drug as a treatment option, showing conflicting results. One study by Butler et al. (2023) involved 25,000 non-hospitalized patients with early-stage COVID-19 who were started on molnupavir. This treatment did not reduce the risk of hospitalization or death; however, there was a reduced time to recovery (9 vs. 15 days) [68]. Other studies showed similar results, suggesting molnupavir is an effective alternative treatment for COVID-19.
3.11. Nirmatrelvir-Ritonavir (Paxlovid)
This is currently the preferred treatment choice against COVID-19. Nirmatrelvir and ritonavir are both protease inhibitors, the latter of which also increases the concentration of the former to target therapeutic range due to its additional inhibition of cytochrome P-40. This combination drug functions by blocking the processing of the viral polypeptides, halting the replication of new virion particles. This mechanism of action is similar to that of lopinavir-ritonavir, however nirmatrelvir-ritonavir differs by targeting a protease specific to SARS-CoV-2. (Fig. 5). Many studies have been performed on its efficacy, demonstrating the following results:
- Hammond et al. (2022) conducted a randomized controlled trial with 2246 patients, where 1120 were assigned to the treatment group and 1126 to the placebo group. The results of the study demonstrated that treatment with nirmatrelvir-ritonavir resulted in a risk of progression to severe COVID-19 that was 89% lower than the risk from the placebo [69].
- Schwartz et al. (2023) conducted a population-based cohort study of adults who had a positive COVID-19 test between April 4th-August 31st, 2022, comparing patients who were treated with nirmatrelvir-ritonavir and those who were not. Of the 177,545 patients in the final cohort, even though only 5% (8,876) were treated with nirmatrelvir-ritonavir, there were still significantly reduced odds among these patients of hospital admission and death from COVID-19 [70].
- Dryden-Peterson et al. (2023) conducted a population-based cohort study to assess whether nirmatrelvir-ritonavir reduces the risk of hospitalization and death among outpatients with early-stage COVID-19. A total of 44,551 patients were included: 12,541 who received nirmatrelvir-ritonavir and 32,010 who did not. While the overall risk was already significantly low (1%) in this population, treatment with nirmatrelvir-ritonavir was shown to reduce this risk even further [71].
While this treatment has proven effective, there are significant drug interactions that may require dose adjustment or avoidance. Special care must also be taken in patients with kidney impairment, as this can lead to toxic levels of the drug.
Many pharmaceutical companies worldwide have also developed vaccines to counter the SARS-CoV-2 virus. Specifically in the United States, the three main developed vaccines include Pfizer-BioNTech (BNT162b2 or Comirnaty), Moderna (mRNA-1273 or Spikevax), and Janssen (also referred to as Johnson & Johnson; Ad26.COV2.S or Jcovden). The first two are mRNA vaccines, while the third one is an adenoviral vector vaccine. BNT162b2 is indicated for individuals 5 years or older, while mRNA-1273 and Ad26.COV2.S are indicated for individuals 18 years old and older [72, 73].
The mRNA vaccines are more favourable over Ad26.COV2.S due to the preferred risk-benefit ratio associated with the mRNA vaccines. Ad26.COV2.S has been associated with thrombosis, thrombocytopenia, and possible Guillain-Barre syndrome, while the mRNA viruses have only been associated with myocarditis. The benefits outweigh the risks for all the vaccines; however, the risks associated with the mRNA are deemed less severe [74]. There have also been many observational studies that determined vaccine effectiveness is higher from two doses of either mRNA vaccines compared to one dose of Ad26.COV2.S. A case-control study examined many immunocompetent adults who were hospitalized with COVID-19, and it was estimated that effectiveness against COVID-19-related hospitalization was 93% and 88% for mRNA-1273 and BNT162b2, respectively, in comparison to 71% for Ad26.COV2.S [75]. There are numerous differences between the three vaccines in terms of immunogenicity (Table 4), efficacy (Table 5), and safety (Table 6) [76-78].
Many vaccines have been produced outside of the United States across the globe. These include University of Oxford, AstraZeneca, and the Serum Institute of India (ChAdOx1 nCoV-19/AZD1222 or Vaxzevria), Novavax (NVX-CoV2373, Covovax or Nuvaxovid), CanSino Bioloigcs (AD5-nCOV or Convidecia), Gamaleya Institute (Gam-COVID-Vac or Sputnik V), Sinopharm (BBIBP-CorV or Vero Cell), Sinovac (CoronaVac), Bharat Biotech/Indian Council of Medical Research (BBV152 or Covaxin), and Zyduse Cadila (ZyCoV-D). Each vaccine has been authorized for usage in at least one country.
3.12. ChAdOx1 nCoV-19/AZD1222 (Vaxzevria)
This vaccine has a two-dose regimen and is based on a replication-incompetent chimpanzee adenovirus vector that expresses the spike protein. Its vaccine efficacy after the second dose is 70-76% (95% CI 54.8-80.6) at 14 days and onwards, although it is suspected to wane over time. It was proven ineffective against the Delta and Omicron variants. Fatigue, headache, and fever were common side effects after receiving the vaccine and were severe in up to 8% of recipients [79-90].
3.13. NVX-CoV2373 (Covovax or Nuvaxovid)
This vaccine is a recombinant protein nanoparticle vaccine comprising trimeric spike glycoproteins and a Matrix-M1 adjuvant. In the United States and Mexico clinical trials, efficacy was found to be 90.4% (95% CI 82.9-94.6) in preventing symptomatic COVID-19 in individuals aged 18-84 years who have never been infected. A clinical trial in the United Kingdom found a lower but nonetheless significant efficacy (82.7%, 95% CI 73.3-88.8) [91, 92].
Table 4.
| - | BNT162b2 (Comirnaty) | mRNA-1273 (Spikevax) | Ad26.COV2.S (Jcovden) |
|---|---|---|---|
| Immunogenicity | • Demonstrated binding and neutralizing antibody response comparable to those in convalescent plasma of individuals aged 18-85 years who had an asymptomatic or moderate COVID-19 infection • Antibodies induced in individuals aged 12-15 years and 5-11 years (with a lower dose) were higher than in those aged 16-25 years • Neutralizing antibodies levels generated are lower against Delta, and even lower against Omicron |
• Demonstrated binding and neutralizing antibody response comparable to those seen in convalescent plasma with vaccination in healthy individuals aged 18-55 years • Associated with higher antibody titers after the second dose compared to BNT162b2 • Neutralizing antibodies levels generated are lower against Delta, and even lower against Omicron |
• Demonstrated binding and neutralizing antibody response in individuals aged 18-85 years after single dose, although slightly lower than those in convalescent plasma • Second dose evaluated in a few participants showed an increase in levels • Neutralizing activity retained against Delta, but are significantly lower against Omicron |
| - | BNT162b2 (Comirnaty) | mRNA-1273 (Spikevax) | Ad26.COV2.S (Jcovden) |
|---|---|---|---|
| Efficacy | • 95% vaccine efficacy (95% CI 90.3-97.6) in preventing symptomatic COVID-19 at 7 days and onwards following the second dose in individuals aged 16 and older • Individuals aged 65 years and up with comorbidities or obesity observe an efficacy of 91.7% (95% CI 44.2-99.8) • Individuals aged 12-15 years without evidence of prior infection observe an efficacy of 100% (95% CI 75.3-100) • Individuals aged ≤11 years without evidence of prior infection observe an efficacy of a lower vaccine dose of 91% (95% CI 68-98) |
• 94.1% vaccine efficacy (95% CI 89.3-96.8) in preventing symptomatic COVID-19 at 14 days and onwards following the second dose in individuals aged 18 and older • Individuals aged 65 years and up observe an efficacy of 86.4% (95% CI 61.4-95.5) |
• The regular single dose regimen had a 66.9% vaccine efficacy (95% CI 59-73.4) in preventing moderate to severe COVID-19 at 14 days and onwards in individuals aged 18 and older • Vaccine efficacy against severe cases trended higher at 78% and 85% after 14- and 28-days post-vaccination, respectively • The trial of two dose regimen showed preliminary efficacy rates against symptomatic and severe COVID-19 at 75% and 100%, respectively |
| - | BNT162b2 (Comirnaty) | mRNA-1273 (Spikevax) | Ad26.COV2.S (Jcovden) |
|---|---|---|---|
| Safety | • Local and systemic side effects are common, especially after the second dose. Generally mild to moderate symptoms • Reported symptoms include reaction at injection site, usually pain (in 65% after either dose), fatigue (29% after first dose vs. 48% after second dose), headache (25% vs. 40%), myalgias (17% vs. 37%), fever, chills and joint pain (20% after second dose) • Myocarditis and pericarditis have also been reported following receipt of the vaccine |
• Local and systemic side effects are common, especially after the second dose. Generally mild to moderate symptoms • Reported symptoms include reaction at injection site, usually pain (in 74% after first dose vs. 82% after the second), fatigue (33% vs. 60%), headache (27% vs. 53%), myalgias (21% vs. 51%), fever and chills (40% after second dose), and joint pain (32% after second dose) • Myocarditis and pericarditis have also been reported following receipt of the vaccine |
• Local and systemic side effects are common, mostly occurring the first day after vaccination. 76% of vaccine recipient reported at least one reaction, mostly fatigue, pain and headache, and 61% reported at least one reaction at injection site • Others reported anxiety-related symptoms including tachycardia, hyperventilation, light-headedness, and syncope • It has been associated with a specific syndrome of thrombosis with thrombocytopenia, and possible associated with Guillain-Barre syndrome |
3.14. AD5-nCOV (Convidecia):
This vaccine has a single-dose regimen and is based on a replication-incompetent adenovirus 5 vector that expresses the spike protein. It was shown to have a vaccine efficacy of 57.5% (95% CI 39.7-70) for symptomatic infection and 91.7% (95% CI 36.1-99) for severe infection [93].
3.15. Gam-COVID-Vac (Sputnik V)
This vaccine has a double-dose regimen and uses two replication-incompetent adenovirus vectors that express a full-length spike glycoprotein. It has an efficacy of 91.6% (95% CI 85.6-95.2) in preventing symptomatic COVID-19 at the time of the second dose. The most common side effects were local and systemic flu-like reactions [94, 95].
3.16. BBIBP-CorV (Vero Cell):
Two inactivated, whole viruses based on two different SARS-CoV-2 isolates from patients in China, WIV04 and HB02, were selected as potential COVID-19 vaccines. Each had an aluminum hydroxide adjuvant and had a double-dose regimen. Vaccine efficacy was shown to be around 73% (95% CI 58-82) for WIV04 and 78% (95% CI 65-86) for HB02 [96]. The HB02 strain was selected for the later approved BBIBP-CorV vaccine [97].
3.17. CoronaVac
This inactivated COVID-19 vaccine has an aluminum hydroxide adjuvant and a double-dose regimen. Vaccine efficacy from a clinical trial in Turkey was shown to be 83.5% (95% CI 85.4-92.1); however, lower rates were reported from trials in other countries. In an observational study in Chile, effectiveness was estimated to be 70% for preventing COVID-19 and 86-88% for preventing hospitalization or death. Another study in Brazil reported lower effectiveness among adults aged 70 years and older, estimating 47% for preventing COVID-19, 56% for preventing hospitalization, and 61% for preventing death [98-101].
3.18. BBV152 (Covaxin)
This inactivated COVID-19 vaccine has aluminum hydroxide and a toll-like receptor agonist adjuvant with a double-dose regimen. It has been shown to have a vaccine efficacy of 78% (95% CI 65-86) against symptomatic COVID-19. There have been no serious adverse events related to the vaccine except for one possible case of immune thrombocytopenic purpura [102].
3.19. ZyCoV-D
This is the first DNA COVID-19 vaccine. It is delivered through a needle-free device with a high-pressure stream. A clinical trial among 28,000 participants showed that efficacy against symptomatic COVID-19 was reportedly 67% following three doses; however, the trial details required for critical review of these results have not been made public [103].
The effectiveness of the COVID-19 vaccines has been demonstrated through multiple clinical trials and published reports. However, concerns have risen over the health risks associated with the use of synthetic mRNA in some of their development [104, 105]. Many studies have stated that the injection of a vaccine containing synthetic mRNA has no potential risk of integration into the host genome [106-109]. This is due to the fact that in cells, transcribed mRNA is transported across the nuclear membrane to the cytoplasm for translation into proteins. This transportation is facilitated by nuclear pore complexes, which are proteins embedded in the membrane that regulate the transportation of molecules in and out of the nucleus. It is this selective function that prevents mRNA from re-entering the nucleus once translation is complete and is the basis for the claims made concerning the safety of mRNA vaccines. Furthermore, the integration of mRNA into the host genome would also require reverse transcription to create complementary or cDNA. This process requires a reverse transcriptase, a DNA polymerase enzyme, which is not normally present in healthy somatic cells.
Other than retroviruses, such as the Human deficiency virus (HIV), the genome from viruses does not typically integrate into the infected host genome. However, documented cases have been reported. In the human genome, roughly 17% encodes for Long Interspersed Element-1 (LINE1) elements, which are a type of autonomous retrotransposons. In somatic cells, LINE1 is normally repressed as it can contribute to the development of disease and cancers. However, de-repression can be seen in ageing cells, cancerous tissues, or virus-infected cells [110]. In the case of COVID-19 infection, studies have found evidence that large fractions of the viral RNA could be reversed, transcribed, and integrated into the genome of the infected cells, later being expressed as viral-host chimeric transcripts. This was achieved through the activation of LINE1 elements, acting as the reverse transcriptase, and was hypothesized to contribute to PCR-positive results being observed weeks after the initial infection. However, as only fractions of the genetic sequences were integrated into the host genome, full viral replication was prevented [111-112].
There have been limited studies examining the risks surrounding the genomic integration of vaccine-injected mRNA. One study in 2022 examined mRNA integration in vitro in a human liver cell line, Huh7, after vaccination with the Pfizer BNT162b2 vaccine. The results demonstrated increased LINE1 activity with significant mRNA reverse transcription into cDNA [113]. However, a few points are important to note. Firstly, the cell line used for the study was a cancerous liver cell line. LINE1 activity is known to be increased in cancerous tissues, which limits the accurate representation of the general, healthy population. In fact, another study that observed SARS-CoV-2 genetic integration into the host genome of infected cells reported no integration when the viral mRNA was transfected into healthy cells. This is attributed to the increased LINE1 activation in the infected cells that would not be seen in the transfected ones [112]. Additionally, the paper did not identify positive host genome integration of the cDNA, only commenting on the fast uptake of the vaccine in the cells and intracellular reverse transcription. Further investigations are required to explore the risk for vaccine-delivered mRNA integration into the host genome in the general population and the potential impact and health risks.
4. VIRUS VARIANTS, GENETICS, AND HOST GENETIC IMPLICATION IN INFECTION AND DISEASE SEVERITY
COVID-19 disease severity has shown to be highly polygenic, with several studies linking multiple genetic variants with levels of severity. The first SARS-CoV-2 strain was detected in Wuhan, China, in December 2019 and was called the L strain. By the end of 2020, the L strain had gone through multiple mutations, producing the S, V, and G strains. Throughout the pandemic, multiple variants of the original strain were identified and are described in Table 7. The five main variants that were monitored included Alpha, Beta, Delta, Gamma, and Omicron, with eight additional lesser monitored variants [114-117]. The most severe variant was the Delta variant, which was believed to be one of the most transmissible of the variants and led to more severe illness and hospitalizations. This was followed by the Alpha variant, which was determined to be more contagious than the original strain but with a lower severity of illness in comparison to the other variants. The Beta and Gamma variants were also believed to be more transmissible than the original strain, and they had an increased ability to evade immune responses. However, as with the Alpha variant, the disease course was generally less severe. Finally, the Omicron variant was considered to have the least severe disease course with decreased hospitalizations and mortality compared to the other variants. Since its detection, it has been divided into multiple subvariants. In terms of vaccination efficacy per variant type, the developed vaccines had the highest efficacy against the original SARS-CoV-2 variants. However, additional vaccine boosters were shown to provide increased immunity for the newer variants, although these effects might only be temporary [118-121].
By early 2023, the COVID-19-associated death count in the United States alone was over 1.1 million. The federal COVID-19 Public Health Emergency (PHE) declaration ended soon after on May 11, 2023, marking the official end of the pandemic. Currently, only the Omicron subvariants are variants of concern (VOC) that are still in circulation.
As there is variation in the severity of COVID-19 symptoms, one study sought to examine the molecular mechanisms that influences the severity. Results identified an association between the risk of severe disease and a multigene locus on chromosome 3p21.31 and the ABO blood group locus on chromosome 9q34.2 [122]. The HLA region did not show any association signal. Individuals with blood group A were determined to have an increased risk, while those with blood group O had a decreased risk. A strong associative signal was observed at the rs11385942 insertion-deletion GA or G variant at the locus on chromosome 3p21.31. The GA allele was observed to have a higher frequency among patients on mechanical ventilation than those who were only receiving supplemental oxygen. This could suggest that this risk allele causes a predisposition to the severe forms of COVID-19. There was no elevated risk-allele frequency observed for the ABO locus [123, 124].
The gene cluster on chromosome 3p21.31 has been identified as the major genetic risk locus for severe symptoms following COVID-19 infection. The genomic segment is a 50-kilobase locus and comprises multiple genes, including LZTFL1, SLC6A20, FYCO1, CCR9, CXCR6, and XCR1. This risk haplotype is inherited from Neanderthals and is carried by around 50% of people in South Asia, around 16% of people in Europe, and 9% of admixed American individuals who carry at least one copy. The highest carrier frequency is seen in Bangladesh, where 63% of the population carries at least one copy, and 13% are homozygous for the risk haplotype. It is almost completely absent in the African populations. Multiple susceptibility-related and cytokine regulation-related genes in immune cells have been assessed and described, including a pLI score. This score is the probability of being loss-of-function intolerant, representing how much a single gene can tolerate mutational variants (e.g., frameshift, stop-gain, etc.). A pLI score of 0.9 or greater indicates the gene can be highly constrained, with mutational variants not frequently observed. The genes with pLI scores under 0.9 are listed in Table 8 [125, 126].
The genetic risk segment includes genes of several chemokine receptors, such as CCR9, CXCR6, and XCR1, which play critical roles in immune responses and pathogenesis. They can be classified into several functional groups, including “inflammatory”, “homeostatic”, and “dual function”. Inflammatory chemokines are upregulated until inflammatory conditions are met with leukocyte recruitment to inflamed tissues. Homeostatic chemokines are continuously expressed in lymphoids and other organs, mediating the homeostatic migration of various cell types. Dual-function chemokines overlap in both fields. Unlike inflammatory chemokine receptors, homeostatic and dual-function chemokine receptors are more restricted, only binding with up to two ligands [127].
Regarding the six candidate genes on chromosome 3p21.31, LZTFL1 is one of the most significant with the rs11385942 variant located within it. The LZTFL1 gene encodes a protein involved in protein trafficking and primary cilia. In T lymphocytes, this gene participates in the immunologic synapse with antigen-presenting cells, such as dendritic cells. LZTFL1 encodes Leucine zipper transcription factor-like 1 that is associated with the immune synapse between an antigen-presenting cell or target cell and a lymphocyte, such as a T/B cell or natural killer (NK) cell. Ultimately, it was observed to modulate T-cell activation as well as increase IL-5 levels, which is a pro-inflammatory cytokine involved with the production of eosinophils [128-130].
A genome-wide association study was performed for genetic variations that may identify mechanistic targets for therapeutic treatments against COVID-19. Additional associations have been shown on chromosome 12q24.13 (rs10735079, p=1.65 × 10-8) in a gene cluster encoding antiviral restriction enzyme activators (OAS1, OAS2 and OAS3), on chromosome 19p13.2 (rs2109069, p=2.3 × 10-12) near the gene encoding tyrosine kinase 2 (TYK2), on chromosome 19p13.3 (rs2109069, p=3.98 × 10-12) within the gene encoding dipeptidyl peptidase 9 (DPP9), and on chromosome 21q22.1 (rs2236757, p=4.99 × 10-8) in the interferon receptor gene IFNAR2. These genetic signals relate to key host antiviral defence mechanisms and mediators of inflammatory organ damage in COVID-19, both of which can be targeted for treatment [131].
The IFNAR2 and OAS genes are involved with innate antiviral defences, which are important in early disease. In contrast, DPP9 and TYK2 genes are associated with host-driven inflammatory lung injury, which impacts late, life-threatening COVID-19 infections. These four genes are described below:
4.1. IFNAR2:
The increased expression of this gene was associated with reduced odds of severe COVID-19, which was in line with the beneficial role of type I interferons. This suggests a protective role against the virus. However, exogenous interferon treatment was not observed to reduce mortality in hospitalized patients, suggesting that its beneficial effect may be mediated during the early stages of the infection when the viral load is high [132].
| Variant type | First outbreak | Earliest detection | Associated mutations |
|---|---|---|---|
| Alpha (B1.1.7) | United Kingdom | September 2020 | H69-, V70-, Y144-, L452R, E474K, S494P, A570D, D614G, T716I, S98A, D1118H, K1191N, N501Y, and P681H |
| Beta (B.1.351) | South Africa | May 2020 | L241-, L242-, A243-, P38RL, K417N, E484K, N501Y, L18F, D80A, D215G, and A701V |
| Epsilon (B1.427/B.1.429) | Southern California | July 2020 | D1183Y, S131, W152C and L452R |
| Delta (B.1.617.2) | India | October 2020 | E156-, F157-, A222V, W258L, K417N, T19R, V70F, T95I, G142D, L452R, T478K, P681R, R158G, D614G, and D950N |
| Gamma (B.1.1.28.1) | Brazil | November 2020 | L18F, T20N, P26S, D138Y, R190S, D614G, H655Y, P681H, T10271, K417T, E484K, and N501Y |
| Kappa (B.1.617.1) | India | December 2020 | E154K, L452R, E484Q, and P681R |
| Lambda (C.37) | Peru | December 2020 | G75V, T761, L452Q, F490S, D614G, and T859N |
| Eta (B.1.525) | UK and Nigeria | December 2020 | A67V, H69-, V70-, Y144-, E484K, D614G, Q677H, and F888L |
| Mu (B.1.621/B.1.621.1) | Colombia | January 2021 | T95I, R346K, E484K, N501Y, D614G, P681H, D950N, and N1074K |
| Iota (B.1.526) | New York City | February 2021 | Y144-, L5F, D80G, T95I, F157S, D253G, L452R, S477N, E484K, D164G, A701V, T859N, D950H, and Q957R |
| Zeta (P.2) | Brazil | February 2021 | E484K, D614G, N501Y, D614G, and P681H |
| Theta (P.3) | The Philippines | February 2021 | E484K, D614G, N501Y, D614G, and P681H |
| Omicron (B.1.529) | South Africa | November 2021 | A67V, T95I, G142D, Y145D, N211I, L2121, G339D, R346K, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, Q496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F |
| Signal | Gene name | pLI |
|---|---|---|
| Intermediate | PLC-ɣ | 0 |
| IRAK4 | 0 | |
| JAK2 | 0.65 | |
| PIK3CG | 0 | |
| GM-CSF | 0.83 | |
| LZTFL1 | 0.06 | |
| SLC6A20 | 0 | |
| FYCO1 | 0 | |
| Chemokine | CCL2 | 0.608 |
| CCL7 | 0.001 | |
| CCR2 | 0.02 | |
| CCR5 | 0 | |
| CCR9 | 0 | |
| CXCR6 | 0 | |
| XCR1 | 0.02 | |
| CXCL10 | 0.37 | |
| Cytokine storm | IL-1β | 0.13 |
| IL-6 | 0.32 | |
| IL-8 | 0 | |
| IL-10 | 0.01 | |
| IL-18 | 0.03 | |
| IL-1RA | 0.03 | |
| TNF | 0.803 | |
| IFN-ɣ | 0.472 |
4.2. OAS Gene Cluster
These genes encode enzymes that produce 2’5’-oligoadenylate (2-5A), which is a host antiviral mediator. Its role is to trigger an effector enzyme, RNase L, which degrades double-stranded RNA, a replication intermediate of SARS-CoV-2 and other coronaviruses. This offers a potential therapeutic treatment, as endogenous phosphodiesterase 12 (PDE-12) degrades 2-5A in the body. The use of PDE-12 inhibitors can, therefore, be used to augment OAS-mediated antiviral activity [133].
4.3. DPP9
This gene encodes a serine protease that is involved with many intracellular functions, including antiviral activities. Some examples are the cleavage of CXCL10, an antiviral signalling mediator, as well as having key roles in antigen presentation and inflammasome activation [134].
4.4. TYK2
This gene is a member of the Janus family and is involved with cytokine signaling and antiviral immunity. It has been chosen as one of four gene targets for JAK inhibitors, such as baricitinib, due to the association between TYK2 expression and critical illnesses [135].
Other important genes include SLC6A20, which is expressed in the gastrointestinal tract and encodes sodium–imino acid (proline) transporter 1 (SIT1), reportedly regulated by ACE2 receptors. This gives evidence as to why individuals infected with COVID-19 infections can experience gastrointestinal symptoms, such as nausea, vomiting, and diarrhea [136, 137]. FYCO1 encodes the FYVE and coiled-coil domain containing 1, which is an autophagy adaptor protein. Studies have found associations between this region and clinical characteristics found in individuals with severe COVID-19 infections. This is due to the likelihood of it being a key mediator linking the primary site of coronavirus replication, ER-derived double-membrane vesicles, with the microtubule network in the host. This offers a potential target for therapeutic treatment against COVID-19, as downregulating expression of FYCO1 could potentially decrease replication and infectivity of the virus [138].
Many studies were performed at Mount Sinai Health System in New York City regarding the molecular profile of COVID-19. One study sought to explore the genetic variations seen in children presenting with multisystem inflammatory syndrome following a diagnosis of COVID-19 [139]. Multisystem inflammatory syndrome in children (MIS-C) presents with symptoms including fever, inflammation, and widespread pathology in multiple organs, similar to those seen in Kawasaki disease [140, 141]. The study involved RNA sequencing of 30 blood specimens obtained from MIS-C patients, pediatric COVID-19 cases, and healthy control genes while implementing different statistical approaches including co-expression and Bayesian probabilistic-causal networks to identify disease-associated genes. Among the MIS-C population, the results showed that there was a decrease in NK cells, cytotoxic (CD8+) T-cells, and a downregulated module of genes associated with mature CD8+ (Tex) cells and CD56dimCD57+ NK cells. CD8+ T cells are capable of regulating one another when confronted with a viral infection, and a depletion of circulating NK cells can lead to CD8+ T cell exhaustion. This exhaustion causes specific CD8+ T cells to have poor cytokine output, cytolytic activity, and inhibition of proliferative capacity. This can lead to severe T cell immunopathology following a viral infection when, on the contrary, its presence can improve the symptoms [142, 143]. Nine key regulators were implicated in these findings, including TGFBR3, TBX21, C1ORF21, S1PR5, PRF1, MYBL1, KLRD1, SH2D1B, and GZMA. All of these genes have known associations with NK cells and CD8+ T cells, as well as illnesses similar to MIS-C [144, 145]. The conclusion indicated that an aberration in these aforementioned genes could lead to a decrease in NK cells with an accompanied lack in CD8+ T cell exhaustion, which may lead to severe inflammatory disease similar to MIS-C. TBX21 has also shown to be the most promising therapeutic target of the nine genes due to its coding T-bet, which is a biological switch in the transition of Texprog2 to Texint during Tex differentiation [146]. However, future studies would be needed to determine why the resolution of a COVID-19 infection would lead to the development of MIS-C.
Another study conducted at Mount Sinai Health System evaluated the role of differing genes among the SARS-CoV-2 variants when developing rapid diagnostic tests. As new variants emerged as the pandemic progressed, there was an increased risk for “target dropout” due to the insufficient amplification of the target, leading to false negatives caused by primer/probe binding site (PBS) mismatches. One diagnostic test is the Agena MassARRAY® SARS-CoV-2 Panel, which utilizes probes for five targets across the N and ORF1ab genes, allowing for a broad platform to accommodate for PBS mismatches. The study used this diagnostic test to determine target results and sequence data for 1262 positive cases of COVID-19. Not surprisingly, the PBS mismatches were higher in specimens with target dropout. It was also observed that among specimens with N3 target dropout, 57% contained an A28095T substitution that is very specific for the Alpha variant. Furthermore, redundancy in target design can be beneficial in preventing false negatives, and understanding target performance can help explain the activity of the numerous SARS-CoV-2 variants [147].
There was limited information on COVID-19 when it was introduced to the United States, as most data was concerned about its spread. Respiratory pathogen-negative nasopharyngeal specimens from 3,040 patients were obtained from the Mount Sinai Health System during the first ten weeks of 2020 for a study attempting to better understand the viral origin and its transmission. These samples were negative for diagnostic molecular amplification testing for routine respiratory pathogens. They were evaluated for the presence of two COVID-19 viral markers, the SARS-CoV-2 specific ORF1ab genes and the pan-Sarbecovirus envelope E gene. Results from this study indicated that the COVID-19 infection was present in NYC at least 6 to 8 weeks before the first official wave of the pandemic and was overlooked as another type of viral respiratory infection [148].
One other study at Mount Sinai Health System attempted to understand the molecular etiology behind the post-acute sequelae of the COVID-19 infection. Blood samples were obtained for whole blood RNA sequencing and serology from 567 patients who were being followed into the post-acute period. The study sought to determine if the relationship between acute phase COVID-19 and the development of post-acute sequelae could be observed in blood gene expression. In addition to the RNA sequencing, cell-type specific changes in gene expression were assessed for association with commonly observed symptoms of the infection. Results showed at least two etiologies for different sets of symptoms, one dependent and one independent of the antibody response. Processes leading to post-acute sequelae had already begun during patient hospitalization, indicating that study designs often missed an important time period to explore the pathogenesis of post-acute sequelae. Additionally, the molecular processes leading to post-acute sequelae were also not simply explained by acute severity. In regard to inflammatory cell response, plasma cells were found to play a key role in this process. The downregulation of genes associated with antibody production that also play a functioning role in pulmonary symptoms were independent of the antibody titers, whereas the upregulation of genes involved in the same processes causing other COVID-19 symptoms (i.e., sleep problems, nausea, skin rash, etc.) were highly dependent on the antibody response. The latter suggests these symptoms are linked to the immune system's response to the virus [149].
A final study at Mount Sinai Health System was performed using autopsy tissues to examine the molecular profiling of COVID-19-induced damage further. Rapid autopsies were performed on two deceased patients with variable medical histories but who passed with the COVID-19 infection. Tissue samples were taken from infected and non-infected areas for comparison using multiscale, next-generation RNA-sequencing methods, which revealed four major regulatory pathways involved in the disease process. These pathways involved blood vessel development, cytokine production, cell activation, and structure degradation. Effectors found within these pathways could provide potential diagnostic and therapeutic targets, including the complement receptor C3AR1, calcitonin receptor-like receptors, (CLR) and the cellular matrix proteoglycan decorin. While this study had a limited sample size, the findings encourage further development of advanced molecular techniques that can be applied to a larger sample size and expand on the understanding of COVID-19 pathophysiology [150].
5. POLYGENIC RISK SCORES (PRS) WITH COVID-19
Polygenic risk scores (PRS) have been widely applied in clinical studies investigating genetic variants associated with complex diseases that have a polygenic architecture. They are particularly useful in discerning the association between genetic scores and disease status, especially in cohorts where there is a higher prior probability of disease. For example, they can be used to assist in diagnosis or to inform treatment choices. A polygenic risk score is calculated by computing the sum of risk alleles per individual, weighted by the risk allele effect sizes as estimated by a genome-wide association study (GWAS) on the phenotype. It is described as a single-value estimate of an individual’s genetic liability to a phenotype. It can be calculated as the sum of their genome-wide genotypes and weighted by corresponding genotype effect size estimates derived from the GWAS summary statistic data (Fig. 11) [151, 152].
Polygenic risk scoring is one approach to associating the risk of severe outcomes in genome-wide association studies that have clearly shown that common complex disorders have a polygenic architecture. This has enabled researchers to identify genetic variants associated with diseases. The variety of host genetic variants that determine susceptibility to COVID-19 infection and severity can be combined in a PRS. Studies have been conducted to compute genetic risk scores in different populations to determine high-risk groups. One study evaluated how well a genetic risk score based on chromosomal-scale length variation and machine-learning classification algorithms can predict the severity of a COVID-19 infection. Three groups of patients with severe COVID-19 infections were selected, one including patients less than 90 years old, the second less than 80 years old, and the third less than 70 years old. These patients were compared against age-matched people deemed normal (i.e., not infected). A significant difference was observed between the three age groups and their corresponding controls. This indicates that the germ line genetics of the infected patients is correlated with COVID-19 severity. However, while it was shown that genetics play a significant role, it is still too early to develop a useful genetic test to predict severity due to limited data [153, 154].
Another study analyzed PRS and history of chronic obstructive pulmonary disease (COPD) in patients and their association with severe COVID-19 disease. The PRS for the participants was calculated using 112 single nucleotide polymorphisms that were believed to be related to COVID-19 disease severity. The history of COPD was factored into the overall risk. The study then used logistic regression models to examine associations of genetic risk with or without COPD and the severity of COVID-19 disease. About 712 of the 430,582 participants in the study developed severe COVID-19, and 19.8% of these patients had pre-existing COPD. When examining the PRS, those with lower risk scores were observed to have a lower chance of developing severe COVID-19 when compared with others who had intermediate or high PRS [155].
A third study examined the causality of specific coagulation factors, such as D-dimers, prothrombin time (PT), von Willebrand factor (VWF), platelet count, and fibrinogen in the incidence of COVID-19 severity. To investigate the causal relationships, SNPs associated with 12 coagulation factors were selected, including VWF, ADAMTS13, tPA, FX, PAI-1, D-dimer, FVII, FVIII, FXI, aPTT, ETP, and PT. Results showed that genetic predisposition to the antigen level of VWF and the activity level of its cleaving protease, ADAMTS13, were causally associated with the incidence of COVID-19 severity. However, their effects were displayed in opposite directions, with plasma VWF antigen level showing a positive association while ADAMTS13 displayed a negative association. Furthermore, the statistical significance of ADAMTS13 could not always be reproduced with repeated tests, which weakens the association. The predictive ability of PRS derived from the VWF-associated genetic variants was then explored together with other critical risk factors, including age, sex, body mass index (BMI), coronary artery disease (CAD), systolic blood pressure (SBP), type 2 diabetes mellitus (T2DM), and COPD. It was determined that PRS of VWF was an independent risk factor in distinguishing severe COVID-19 infections from healthy controls, with an observed 16% higher risk [156].
Furthermore, when investigating the contribution of VWF PRS in relation to the other critical risk factors, age was determined to be the most important risk factor for COVID-19 severity. However, VWF PRS showed a larger normalized effect size than SBP, which emphasizes that while it is not the most important factor, its predictive value is still significant in determining prevention and a personalized treatment plan. Therefore, monitoring plasma levels of VWF can assist with developing strategies to control severity as well as associated thrombotic complications [156].
CONCLUSION
The COVID-19 disease has been shown to have significant complexity, eliciting symptoms of differing severity from person to person. This increased complexity led to the development of multiple different treatment types, all with varying levels of success. The previously performed studies surrounding the causative virus SARS-CoV-2 have shown that while further research is required, exploring the molecular makeup across different populations can assist in better understanding pre-existing risks for severe COVID-19 infections and their prognosis. This can further help with developing treatment and targeted therapies, combat the disease more efficiently, and continue to help move past this pandemic worldwide.
AUTHORS' CONTRIBUTIONS
It is hereby acknowledged that all authors have accepted responsibility for the manuscript's content and consented to its submission. They have meticulously reviewed all results and unanimously approved the final version of the manuscript.