All published articles of this journal are available on ScienceDirect.
Study of Various Diagnostic Tests for COVID-19: A Review
Abstract
The coronavirus disease of 2019 (COVID-19), a nightmare of this century, has become an ongoing global health emergency for the entire world. This dreadful disease is believed to have originated from China and has now spread worldwide. To date, more than 170 million people have been found affected by this virus, namely “severe acute respiratory syndrome coronavirus-2” (SARS-CoV-2). With the exponential increase in the patients affected by the SARS-CoV-2, the need for testing has also increased tremendously. Early diagnosis is essential to prevent the extensive spread of the disease because of the faster rate of infection. In this regard, various diagnostic techniques are employed for the detection of the infection in symptomatic and asymptomatic COVID-19 individuals. To provide diagnostic care for the control of the disease, various tests like serological testing, nucleic acid amplification test (NAAT), rapid antigen-based testing, and paper-based testing have been developed and are presently in good use. The present mini-review is an attempt to outline the currently available diagnostic kits for the detection of the SARS-CoV-2 causing COVID-19.
1. INTRODUCTION
Coronaviruses are single-stranded RNA viruses. These viruses cause the common cold and other severe acute respiratory syndromes (SARS) in humans. Human coronaviruses are dated back to 2003 when SARS-CoV was witnessed. Subsequently, in 2012, MERS-CoV (the Middle East Respiratory Syndrome coronavirus) became the reason for another pandemic in Saudi Arabia. (Middle East respiratory syndrome coronavirus (MERS-CoV) – The Kingdom of Saudi Arabia, 2020). In December 2019, a more virulent strain was first detected in Wuhan city, China, where patients were first brought in for severe pneumonia-like symptoms. From China, the virus spread to other parts of the world very quickly, and by February 2020, it was declared a pandemic by WHO. Because of the large number of positive cases, the health system of even the developed countries was under tremendous pressure, and to prevent the spread of the disease, countries went into lockdown, resulting in stagnation of industrial and commercial activity worldwide
To date, 171 million confirmed cases and 3 million deaths have been reported as per the worldometer. Even after more than a year of the spread of this viral pandemic, the proper medication for the complete cure of the COVID-19 is yet to be reported.
Early diagnosis is essential for proper control and management of the pandemic. One major problem against this diagnosis is the presence of nonspecific symptoms, like dry cough, headache, sputum production, vomiting, nausea, hemoptysis, fever, dyspnea, myalgia, fatigue, and diarrhea. Loss of smell and taste was found to be early markers of the disease [1]. It can cause acute respiratory distress syndrome and organ failure in the elderly and people with chronic respiratory diseases, which is a possible cause of the current death rates [2].
After the infection, humans can remain symptomatic as well as asymptomatic. Asymptomatic individuals, though found to be less infectious, can release large numbers of this virus, thus becoming effective and untraceable transmitters. The virus is transmitted through droplets, aerosol and sneezing and the incubation period for COVID-19 ranges from 3-14 days [1].
Hence, the improvement of current diagnostic measures to increase detection of early and asymptomatic cases and removal of false positives and negatives are the need of the hour.
In this review, we have tried to discuss various clinical tests used for the detection of the virus with comparable sensitivity and specificity (Fig. 1).

2. REAL-TIME RT- PCR TEST
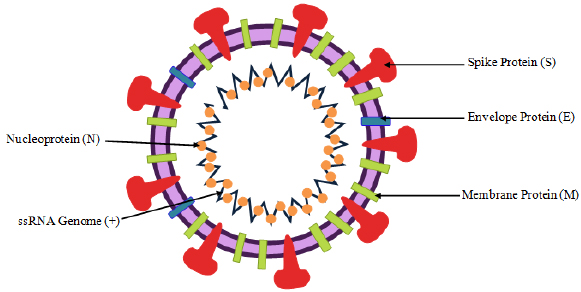
REAL-TIME RT- PCR test (Reverse Transcriptase- Polymerase Chain Reaction) was the first method developed for COVID-19 detection and is considered to be the gold standard test among the available Coronavirus tests. The coronavirus pandemic (COVID-19) is caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). SARS-CoV-2 is an enveloped virus with a genome of single-stranded positive-sense RNA (ssRNA) [3]. The SARS-CoV-2 has structural, nonstructural, and accessory proteins, as shown in Fig. (2). Structural proteins include the S protein or the Spike protein encoded by gene ORF2. It is said to have 2 subunits - S1 and S2. S1 binds to the host receptor, and S2 mediates host and viral membrane fusion. The M protein or the Membrane protein is coded by gene ORF5. It is the most abundant of all the viral proteins and shows interaction with the N protein of the virus to enable RNA packaging. E protein or Envelope protein, coded by gene ORF4, is a small membrane protein and ensures viral assembly and parthenogenesis. The N protein or Nucleocapsid protein is coded by ORF9a. It shows interaction with M protein and non-structural protein 3 (Nsp3) to aid in viral replication and other processors involving invasion in the host immune system [4-7].
The identification and sequencing of the SARS-CoV-2 genome have enabled the development of an RT– PCR test. The RT- PCR involves the use of reverse transcription of SARS-CoV-2 RNA into cDNA (Complementary DNA) strands. This is followed by amplification and detection of cDNA at specific regions. These specific regions can be used to detect the SARS-CoV-2 from any other SARS-CoV virus. The amplified DNA is then detected in real-time using fluorescence probes. The fluorescence intensity reflects real-time amplification of the DNA sequence and is used for quantitative detection of the target DNA [8].
There are different targets present to detect SARS-CoV-2. These include genes encoding for the N, S, and E protein, the open reading frame 1 ab (Orf1ab), and the RNA Dependent RNA Polymerase (RdRP) gene that is located within (Orf1ab). The gene E is highly conserved among all the beta coronaviruses, while the N gene may cross-react with other coronaviruses. The RdRP gene can also be used in differentiating between the SARS-CoV-2 and other Coronavirus. Gene S is also very useful in differentiating SARS-CoV-2 because it is highly divergent from other coronaviruses [9].
During the time of the outbreak, seven RT-PCR assays were developed by scientists from all over the globe for the diagnosis of COVID-19. These different protocols provide access to different targeting genes. One of the major challenges with this detection technique is the false-negative result of actual COVID-19 patients. Several factors contribute to false-negative results. These may include improper sample collection, inefficient removal of the sample matrix, impurities loss or degradation of the target RNA molecule, and inadequate purification of RNA.
Therefore, a positive RT-PCR is indicative of active infection with SARS-CoV-2, but a negative result may not be correct. Thus, the result of real-time RT-PCR must be cautiously interpreted [10].
3. NAAT- NUCLEIC ACID AMPLIFICATION TESTING
NAAT test is based on the principle of testing the viral RNA present in the body of an infected person and can be performed using the Reverse Transcriptase Real-time PCR technique to carry out the amplification of the Viral RNA. NAAT detects the viral RNA by binding the primer to its nucleic acid chain [11]. It also detects RdRp, nucleocapsid, envelope, and spike protein gene of the virus to give a positive test result [12].
The test is highly sensitive and provides highly specific results. Both nasopharyngeal (NP) swab and oropharyngeal (OP) swab samples can be used to carry out the test, but it is preferred to collect both upper and lower track respiratory samples to get better results [12, 13].
It has been found that nasopharyngeal samples are preferred over oropharyngeal samples for the swab-based testing as the false negative percentages were 8.4% and 10% for NP swab and OP swab, respectively [12]. Since the test is highly sensitive and specific, so even small alterations in the testing process, like poor specimen quality, inadequate viral load, viral mutation at the time of sampling, etc., might cause incomplete or false test results. It should be noted that a negative result does not rule out the possibility of a SARS-CoV-2 infection as it also depends on the time of collecting the sample because the symptoms of the disease come after a time duration of about a week to 15 days [12, 13].
4. SEROLOGICAL TESTING
Serological assays measure the response of antibodies to pathogens in bodily fluids, especially blood serum or plasma, in response to SARS-CoV-2. These tests took time to develop because of the sensitivity and specificity required to distinguish between the immunity imparted by COVID-19 and other coronaviruses. The assay uses different platforms, like the lateral flow assay, enzyme-linked immunosorbent assay (ELISA), or protein microarray [14].
4.1. Lateral Flow Immunoassay (LFIA) or Immunochromatographic Test
This method utilizes the property of antibodies to selectively bind to specific antigens. Whole human blood, plasma, stool, sweat, tears, and other fluids are assayed and the presence of a particular molecule is determined. It is based on the biochemical interaction of antigen-antibody or probe DNA-target DNA hybridization. The technique is user-friendly, cost-effective, and has a short assay time as the result can be obtained within 30 minutes [15, 16].
A lateral flow assay (LFA) device has 4 parts, as shown in Fig. (3).
1. Sample pad (area of loading the sample for testing).
2. Conjugate pad (labeled tags are combined on this part).
3. Reaction membrane with a test line and a control line (for assessing the interaction between antigen and antibody).
4. Absorbent pad (meant for reserving waste).
4.1.1. Working
The sample, to be tested, is run along the surface of a pad with reactive molecules that indicate a visual positive or negative result. These pads have a series of capillary beds made of porous paper or a sintered or microstructured polymer. The entire strip is backed by plastic.
Steps for Lateral Flow Assay [17] are as follows:
1. Antibody development against target analyte.
2. Preparation of gold nanoparticles.
3. Preparation of conjugates.
4. To construct a lateral flow strip and construct the analyte.
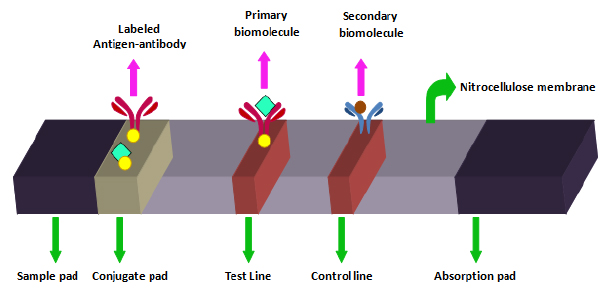
In LFIA, a fluid carrying the analyte moves by capillary action through polymeric strips. These strips contain molecules that can interact with the analyte. A lateral flow test strip is made up of overlapping membranes. The sample is applied to one end of the strip known as the sample pad and it contains suitable buffer and surfactants. The sample then moves to the conjugate release pad that holds antibodies specific to the target analyte and is bound to fluorescent particles, like colloidal gold and latex microspheres. Then, this sample conjugated with antibodies bound to the target analyte migrates to the detection zone, a nitrocellulose membrane with specific biological molecules, like specific antibodies and antigens, that interact with the analyte and give a response on the test line. Control line ensures proper flow of sample. The result lines can be assessed easily by eyes [18].
4.1.2. Lateral Flow Tests in COVID-19
For COVID-19 testing, a specimen taken from the patient (swab from nose or throat) is applied to the lateral flow strip. In case of the presence of SARS-CoV-2 antigen in the extract, it will bind to the SARS-CoV-2 monoclonal antibody. While passing the test line on absorbent paper, the complex is captured by the SARS-CoV-2 antibody, which results in coloring, thus revealing whether the virus is present in the person being tested or not. A control line is to indicate the proper liquid flow through the strip and the result can be assessed by eye or using a dedicated reader.
The sensitivity of the test ranges between 88.75% to 99.17%. It depends on many factors, like the viral incubation period in the host body, viral load in the body, etc., which makes it tough to distinguish between asymptomatic affected and symptomatic affected individuals. The sensitivity of the test is suboptimal, and the risk of false negatives is high when the virus is not very active [15].
4.2. Enzyme-Linked Immunosorbent Assay (ELISA)
This test is more sensitive for the diagnosis of COVID-19. The presence of immunoglobin is detected in this test. The initial humoral response is provided by IgM during the initial stages of the infection, followed by IgG response which is long-term. Antibodies of IgM class can be found in human blood after 6 days of infection while IgG can be detected after 8 days. Therefore, the presence of IgM is an indication of recent infection while IgG presence indicates past exposure, as summarized in Table 1 [15].
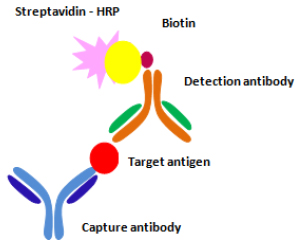
An enzyme-linked immunoassay is used to label antigens and antibodies using enzymes. For this purpose, alkaline phosphates (ALP), β- galactosidase, and horseradish peroxidases (HRP) are used. The antigen is immobilized on the solid phase, like a microtiter plate. The antigen is then allowed to interact with a specific antibody. This interaction is detected by secondary antibodies that are also enzyme-labeled (Fig. 4). This results in the development of color by the use of chemiluminescent substrates, like chloro-5-substituted adamantyl-1,2-dioxetane phosphate used for ALP and luminal for HRP [19].
Generally, these reactions are completed within 30-60 minutes and the reaction can be stopped by the addition of some appropriate solutions, like sodium hydroxide, hydrochloric acid, sulphuric acid sodium azide, and sodium carbonate [19].
4.3. Protein Microarray Method (PMM)
This is a proteomic screening technique for the quantitative and qualitative analyses of a mixture containing many proteins. In this technique, a supporting surface (e.g. modified glass plate, nitrocellulose membrane, or microtitration plate, etc) is laced with ‘capturing proteins’ that are immobilized and capture antibodies or enzymes. The protein analyte can be modified using markers (fluorescence, luminescent, radioisotope markers, etc.) and their interaction with matrix proteins results in an analytical signal [15].
| Phase of Infection | Testing for IgM | Testing for IgG |
|---|---|---|
| Window period | Negative | Negative |
| Early stages of infection | Positive | Negative |
| Active phase of infection | Positive | Positive |
| Late recurrent stages of infection | Negative | Positive |
| Post-infection | Negative | Positive |
| No infection | Negative | Negative |
The serological screening is done for individual peptides present in a viral proteome. SARS-CoV-2 proteome has about 5000 peptides. Its sequence was established from virus isolate 4rdnkr Wuhan-Hu-1 (GenBank ID: MN908947.3), and a PMM test was developed as PEPperCHIP® SARS-CoV-2 Proteome Microarray (PEPperPRINT). The sensitivity of PMM was found to be more than ELISA tests in the case of SARS-CoV 2002 in China. Therefore, this test is more reliable and allows the screening of protein antigens of the virus. There is a possibility of automation in this process. However, among other issues, there is a problem of high-cost equipment that is needed for diagnosis, making PMM less accessible in standard labs [15].
4.3.1. Need for Serological Testing
Serological testing is required to estimate rates of asymptomatic cases and the spread of the disease [20]. Serological data can also provide information regarding re-infection. The antibodies provided must be robust enough to prevent the virus from further attack. Such information is unclear and can help with the development of vaccines and therapeutic drugs. Serum from treated patients can then be utilized and these people can be allowed across borders under ‘immunity passport’ [14].
5. RAPID ANTIGEN DETECTION TEST
This test works on the principle of detecting the viral antigen of SARS-CoV-2. It was developed due to its ease of use and for its rapid result. This test is easy to perform and gives results much faster as compared to other tests used for diagnosis. The main objective of studying this test was to determine its limit of detection (LOD) between RAD test, viral culture, and RT-PCR and its performance [21]. The methods used for testing the performance of the RAD test are as follows:
5.1. Sample Isolation
1. A sample was tested for the culture isolates of SARS-CoV-2, isolated from the strain Human Coronavirus 2019 (hCoV-19). To compare the reactivity and efficacy of this test along with the viral strain, several other viral strains, like Influenza A, adenovirus, and others were also used [21].
2. Samples collected from SARS-CoV-2 confirmed that individuals were isolated and tested for the presence of viral antigen. In another study, both symptomatic as well as asymptomatic patients were tested using the RAD test [21, 22].
5.2. Sample Processing
Two types of samples were isolated, the less viscous and the more viscous. Less viscous samples required no preparation and were directly loaded to the device, while viscous samples were used after dilution [21].
5.3. Principle
The RAD test is solely based on the sandwich principle, this is inspired by the already existing pregnancy tests and based on the color detection principle. The lateral flow of the collected sample is run on a chromatographic material, like nitrocellulose. The areas inside the test kit are pre-administered with antibodies, arranged as monoclonal antibodies and polyclonal antibodies (immobilized), followed by secondary polyclonal antibodies arranged successively. When the sample is run, the nucleocapsid protein of SARS-CoV-2 binds to the regions having monoclonal antibodies or immobilized polyclonal antibodies while the Spike protein of the SARS-CoV-2 is said to bind to the secondary polyclonal antibodies present. In either of the three cases, a color detection will indicate the antigen and antibody binding reaction, which eventually confirms the test. The color change in the area with immobilized polyclonal antibodies is the sole representative of positive or negative test detection [23].
6. RESULTS
It has been reported that the RAD is 1000 times less sensitive than RT-PCR when tested with the BIO CREDIT COVID-19Ag test kit [21]. In another set of experiments with the Bioeasy Biotechnology Antigen kit, the sensitivity of this test was achieved to be 93.7% [24]. The sensitivity of the Panbio Rapid Antigen test was 73.3% and the median threshold obtained was 23.28. It showed that the sensitivity of the detected sample was based on the time of symptom onset and the concentration of the viral sample loaded [22].
In another study utilizing COVID-19Ag Respi-Strip Kit, the sensitivity of the test came down to 50%, since only 47 tests were adequately detected in the sample size of 138 patients, with 94 positive samples [25]. In 16 conducted studies, the sensitivity of the RAD test, compared to the standard RT-PCR test, was 87.8% [26]. While another study involving STANDARD F COVID-19 Ag fluorescent immunoassay (FIA) detection kit depicted that the RAD test is only fit for use within the first few days of the infection [27]. Test performed on a whole blood sample of the fingertip of 3 groups based on the time of infection (0-7 days, 8-15, and >15 days) gave sensitivity values as 18.8%, 100%, and 100%, respectively [28].
7. PAPER-BASED TESTING – FELUDA
This is a novel test developed in India and has been recently approved by the DCGI (Drugs Controller General of India) and is now being used at INR 500 per test [29]. FELUDA stands for “Francisella novicida Cas9 (FnCas9) Editor Linked Uniform Detection Assay,” and is based on CRISPR-Cas9 technology (Clustered Regularly Interspaced Short Palindromic Repeats). The development and application of this cutting-edge technology have earned Emmanuelle Charpentier and Jennifer A. Doudna, the 2020 Nobel Prize in Chemistry. CRISPR-Cas9 proteins can bind to the target DNA or RNA sequence. This binding causes conformational changes in the protein causing the target to cleave. This gives out a signal outcome. A Cas9 ortholog is taken from Francisella novicida, giving the technique its name. The sensitivity of FnCas9 is high and thus, it forms the basis for accurate identification of the target, in this case, SARS-CoV-2 [30].
The method for the collection of swabs is the same as in RT-PCR and Rapid Antigen Testing. Paper-based testing works like a pregnancy test kit and results can be read with the naked eye, where the presence of two lines indicates a positive result. Results can be read within 45 minutes. FELUDA does not require any expensive equipment, making it very feasible. The test has a sensitivity of 96% and a specificity of 98%, which rule out false negatives and false positives [30].
8. LOOP-MEDIATED ISOTHERMAL AMPLIFICATION
Loop-mediated isothermal amplification (LAMP) based protocols enable the efficient amplification of nucleic acids at a single point temperature. This makes it a strong contender for direct field applications, since incorporating the thermal cycling steps in PCR assays has traditionally been a significant limitation for point-of-care devices. This technique works efficiently even with crude sample preparations compared to traditional PCR methods, and also offers a very high amplification efficiency since it is not limited by a doubling-per-cycle threshold [31].
A team led by Di Liu and Jing Yuan reported RT-LAMP assays for SARS-CoV-2 detection, with ORF1ab and S genes as the primer-probe targets [32]. They claim complete detection within 60 min, using a colorimetric detection system that employs fluorescent calcein, where a color change from orange to green indicates a positive reaction.
Another method involving primers targeted at the RdRp utilizes cresol red (a pH-sensitive indicator dye) for the assay readout [33]. Since a proceeding amplification can progressively change the buffer pH from alkaline to acidic, a color change from burgundy to orange/yellow signals a positive reaction.
These innovations in assay readout methodology imply a significant advancement since they make it possible to have even untrained healthcare workers administer, conduct, and interpret diagnostic tests. A noteworthy instance of adapting RT-LAMP for a true bedside point-of-care application is the innovative closed-tube test developed by researchers at the University of Pennsylvania [34]. They combined straightforward sample collection with single or two-step RT-LAMP amplification protocols, along with a visual detection system based on leuco crystal violet (LCV– an intercalating agent which colorimetrically detects double-stranded LAMP amplicons) This device is a readily deployable and highly portable testing method that promises to be a cheap and reliable alternative suitable for all testing environments.
9. NUCLEIC ACID SEQUENCE BASES AMPLIFICATION (NASBA)
A new highly sensitive, affordable, rapid clinical diagnostic technique used to detect viral DNA using fluorogenic RNA aptamer which is based on in vitro isothermal amplification of RNA is known as nucleic acid sequence bases amplification (NASBA) [35, 36]. This assay consists of two sets of primers and three enzymes. The enzymes present in NASBA include a mixture of T-7 ribonucleic acid (RNA) polymerase, reverse transcriptase, and ribonuclease (RNase) H along with a specially designed probe and a pair of specially designed primers [37]. The single-stranded RNA is annealed by primer 1 in the first reaction phase of NASBA, following the synthesis of a complementary DNA (cDNA) strand, and then the formation of the RNA: DNA hybrid takes place. Subsequently, the RNA chain is hydrolyzed by the RNase H and generates the single-stranded DNA. The primer 2 anneals with the reverse transcriptase in the next step and synthesizes double-stranded DNA with the promoter region, recognized by T7 RNA polymerase [37]. Once the ds DNA is formed with the promoter region, the reaction enters into cycles of continuous transcription, reverse transcription, and subsequently the hydrolysis of the RNA which is present in the RNA: DNA hybrid [37]. As each step of transcription can generate nearly 10-1000 RNA copies, the NASBA requires fewer cycles compared to LAMP or PCR. It also results in less overall error frequency and reduced total incubation time. Therefore, as an isothermal amplification process, NASBA is highly specific and suitable for the detection of single-strand RNAs but not for double-stranded DNA.
| S.No | Test | Sample Type / Kit Name | Sample Size | Sensitivity (%) | References |
|---|---|---|---|---|---|
| 1 | Rapid Test | Nasopharyngeal aspirate and throat swab / BIOCREDIT COVID-19Ag test | 47 | 34.3 | [21] |
| Nasopharyngeal swab and throat swab / BIO CREDIT COVID-19Ag test | 51 | 45.7 | |||
| Sputum / BIOCREDIT COVID-19Ag test | 50 | 11.1 | |||
| Throat Saliva / BIOCREDIT COVID-19Ag test | 63 | 40 | |||
| 2 | RAPID TEST | 0-7 days post infection / Bioeasy | 76 | 94.7 | [24] |
| 8-12 days post infection / Bioeasy | 42 | 80 | |||
| 3 | RAPID TEST | Symptomatic Patients / Panbio | 26 | 97.1 | [22] |
| 9 | 77.8 | ||||
| 1 | 30 | ||||
| 1 | 14 | ||||
| 4 | RAPID TEST | Healgen COVID-19IgG/IgM Rapid Test Cassette | NA | 100 | [15] |
| 5 | RAPID TEST | Biomedomics COVID-19IgM- IgG Rapid Test Kit | NA | 96.7 | [15] |
| 6 | RAPID TEST | Phamatech | NA | 86.7 | [15] |
| 7 | Antigen test | <8 Days Post Symptom | 38 | 96.4 | [38] |
| 8 | NAAT | Aptima | 19261 | 99.91 - 99.98 | [12] |
| in-house RT-PCR | 336 | 97.4- 99.1 | |||
| 9 | NAAT | ID NOW | 75 | 85 | [39] |
| Simplexa | 75 | 97 | |||
| m2000 | 75 | 99 | |||
| Xpert | 76 | 98 | |||
| 10 | RT-PCR | RealStar SARS-CoV-2 RT-PCR | NA | 92 | [15] |
| 11 | RT-PCR | ExAmplar COVID-19real-time PCR Kit [L] | NA | 95 | [15] |
| 12 | RT-PCR | AccuPower® SARS-CoV-2 Real-Time PCR Kit | NA | 100 | [15] |
| 13 | RT-PCR | Symptomatic Patients / COVID-19PRESTO and COVID-19-DUO | 381 | 100 | [28] |
| 14 | RT-PCR | <8 Days Post Symptom | 38 | 100 | [38] |
| 15 | ELISA | Euroimmun SARS-CoV-2 ELISA [IgG] | NA | 90 | [15] |
| 16 | SEROLOGICAL TEST | Tianjin Beroni Biotechnology SARS-CoV-2 IgG/IgM Antibody Detection kit | NA | 90 | [15] |
CONCLUSION AND PERSPECTIVE
The rapid spread of COVID-19 is devastating the globe, therefore implementing fast and widespread diagnostic tests is of paramount importance for population screening. The importance of scaling up testing can not be ignored. The diagnostic presentation of available tests varies widely. Initially, the Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) was the diagnostic method of choice, but due to its long waiting time of three to four hours and the dependence of test results on the quantity, type, and timing of specimen collection, this method is not considered very ideal for rapid diagnosis. Additionally, cases of false negatives and false positives can arise, however, these can be avoided to a certain extent by the use of proper techniques and caution. Thus, under any circumstances, ruling out the possibility of infection only based on RT-PCR is not recommended. The use of various NAAT techniques provides a faster method of testing. Although the results for the two tests are comparable, both techniques use a fair amount of expertise and equipment, making them expensive.
Along with PCR tests, serological testing is also very popular and relatively easier to perform. LFIA uses a simple detection method for antibodies and test result is easily read by the naked eye in the form of lines. However, the sensitivity of the test depends on the activity level of the virus and the test can give a false negative in the early stages of infection. ELISA, therefore, is more sensitive for COVID-19 testing. The test can give quick results and detect early stages of infection as well. It also gives a procedure for confirming past infections. PMM showed more sensitivity compared to ELISA and therefore, is now being used for COVID-19 testing as well. These tests, however, play a major role in the population study of the virus. RAD has played a major role in COVID-19 testing, even with its lower sensitivity than that of PCR, mostly due to the rapid results it provides. Also, it does not require sophisticated instrumentation. FELUDA is a more recent technique, developed using CRISPER-Cas9 technology by the scientists of an Indian institute (CSIR-IGIB), and is known for its accuracy. Nevertheless, the test so far has been promising and paves the way to an easier, cheaper, and accurate way of testing for SARS-CoV-2. When compared to the RT PCR test, the FELUDA molecular test just takes 45 minutes to give results after the RNA has been extracted from the patient sample, while the RT PCR test takes about four to five hours of time in the laboratory.
In the wake of an emergency situation surrounding the pandemic caused by SARS-CoV-2, the tests have not yet undergone the vigor that they might with time. Hence, all the tests tend to have shortcomings that are to be overcome shortly. It is the responsibility of the clinical community to understand the test performances and use that information in patient care. The diagnostic tests should be accompanied by standardized, clear, and comprehensible information on performance for clinicians and patients. With the incoming of new information (new strains) about SARS-CoV-2 every day, the development of new tests or modifications of the already existing ones has never stopped. As more and more people get tested for the virus and the increasing studies on its spread, information regarding re-infection can become more clear. Various tests reported so far have been summarized in Table 2.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We would like to thank Prof. Shrikant Kukreti, Department of Chemistry, the University of Delhi, for his valuable suggestions at the time of preparing this manuscript. The authors would also like to thank Prof. Rama, Principal Hansraj College, for her motivation and support to students and teachers.


