All published articles of this journal are available on ScienceDirect.
Impact of SARS-CoV-2 Mutations on Global Travel and the Increasing Number of Re-Infections: A Risk-Assessment Perspective
Abstract
The outbreak of coronavirus disease 2019 (COVID-19) has stunned the world owing to the surreal, unprecedented, and completely unbelievable manner in which it has spread globally within a short span of time. This spread has led to the common combination of variety and has promoted the passage of species blockade and genetic combination of these types of viruses. Despite the short history of the COVID-19 outbreak, with its global spread and frequent mutations, it has impacted the whole world and has become a worrying threat to the society. Scientific reports have disclosed that members of the coronavirus family, such as SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), HCoV-NL63, HCoV-229E, HCoV-OC43, and HKU1 have infected the humans earlier too and that mutations in these viruses have resulted in the more complex severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In the present review, we have discussed how scientists keep track of the genetic tweaks to SARS-CoV-2 as it spreads globally.
Currently, the only way to prevent more such outbreaks is maintaining social distancing, adhering to the World Health Organization guidelines and de-globalizing the world. Genetic variations/mutations reported to date in coronaviruses hint at their cryptic spread.
Scientists are scouring the viral genome for mutations that might reveal how dangerous the pathogen is or how fast it spreads. Cases have been documented in almost all countries, and the mutations in the virus have created problems for the researchers in formulating effective vaccines. Furthermore, global travel has been severely affected after the new mutants have been detected. Therefore, more scientific investigations are necessary to understand how SARS-CoV-2 is likely to mutate in the future.
1. INTRODUCTION
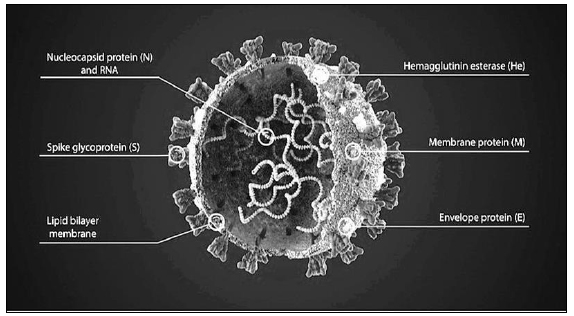
Travelling is the most compelling way to meet new people and explore new places if the route is free of infections, offence, and disasters. For most part of the time, with accurate awareness, the threats can be reduced and the journey memoir is filled with remarkable experiences. Epidemics and pandemics are two of the terrifying news for travelers. In such scenarios, it might be tricky or impracticable to evade the illness. Moreover, not only the travelers but also the other individuals with whom contact is established during the trip are at risk. The pathogens are highly infectious and mutate swiftly. As a result, new and unforeseen outbreaks can happen anytime. Coronaviruses are mostly transmitted from animals to humans. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a well-known respiratory pathogen related to several respiratory illnesses, including pneumonia, common cold, bronchitis, fatigue, and in some cases, kidney failure [1]. SARS-CoV-2 is a single-stranded RNA virus and is 50–400 nm in diameter and comprises four major intrinsic proteins, namely, spike S, envelope E, membrane M, and the nucleocapsid N [2] (Fig. 1).
The S protein enables the virus to attach itself to the membranes of the host cells. The host cells containing angiotensin-converting enzyme-2 (ACE-2) receptors are the main targets of the S proteins. The virus then causes intrinsic alterations to combine with the host cell, finally permitting the viral genetic material to enter the cells [3-5].
As per the Centers for Disease Control and Prevention (CDC), the spread of COVID-19 is typically human-to-human and occurs through respiratory droplets within a distance of 1–2 meters. These viruses may also spread if a person touches the mucosal lining with contaminated hands [6]. Polymerase chain reaction (PCR) is used to identify the viral RNA; in case the test is positive, the presence of SARS-CoV-2 is established. False-negative results are possible and, in symptomatic individuals, additional testing may be necessary [7, 8].
In recent years, the evolution of human coronaviruses has also been accelerated by elements such as urban growth and poultry farming. These activities have allowed the integration of a variety of strains and have facilitated the passage of species hurdle and the genetic combination of these viruses [9]. So far, seven human coronaviruses are known, with SARS-CoV-2 being the latest one. Five of the human coronaviruses have spread internationally and contribute to approximately one-third of widespread cold infections in humans [10]. As reported previously, these viruses can also cause life-threatening bronchiolitis and pneumonia, particularly in children, elderly, and immunocompromised individuals [11, 12]. Furthermore, respiratory illnesses may result in neurological disorders [13-17]. The fifth strain of coronavirus (SARS-CoV) initially emerged in the year 2002 in the Guangdong area of China as uncharacteristic pneumonia with a distinctive headache and fever, followed by respiratory symptoms such as cough and pneumonia, which later developed into serious respiratory failure and acute respiratory distress syndrome [18]. Human coronaviruses, being highly contagious, have rapidly spread across 216 countries and have infected 48.53 million humans, with a death rate of 2.47% [19, 20].
According to Lim et al., for 14 years, the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) and MERS-CoV has propelled COVID-19 into the forefront of research owing to its far above-the-ground contiguity in humans [21]. The study of SARS-CoV-2–host associations has to be aided widely to our lenient COVID-19 development. Human coronaviruses are respiratory pathogens that have been identified to be transmitted to tissue cultures and cell lines acquired from the respiratory tract. Infection of these tissues and cells may induce apoptosis [22, 23]. Human coronaviruses have also been known to infect the immune system and stimulate apoptosis in immune cells such as macrophages, monocytes, T lymphocytes, and dendritic cells [22, 24-28]. COVID-19, named by the International Committee on Taxonomy of Viruses (ICTV) and previously known as “2019 novel coronavirus,” is a pandemic (declared by the World Health Organization (WHO) on 11 March 2020) that is gradually spiraling out of control. The disease is spreading around the world at a very rapid pace. Laboratory facilities are overstretched, and healthcare professionals are insufficient in number to cope with the situation. Patients admitted or to be admitted to intensive care units because of other medical complications will die in large numbers due to the unavailability of hospital staff who are occupied in arresting this pandemic. WHO is monitoring the rate, extent, and trend of global spread. However, presently there is no conclusive evidence that the virus is spread by non-living objects [29]. Doctors and other healthcare professionals treating patients understand that the virus is present in significant quantities in the patients’ upper respiratory tract, thereby causing its spread by sneezing or coughing or by sputtering minute droplets into the air and toward surfaces [30]. According to Kampf et al., coronaviruses can persist on inanimate surfaces, such as papers, steel, gloves, glass, plastic, glass, and wood, for 8–9 days; however, they can be professionally inactivated by surface disinfection measures with 62%–71% C2H5OH (ethanol), 0.5% H2O2 (hydrogen peroxide), or 0.1% NaOCl (sodium hypochlorite) within 60 seconds [31, 32]. The healthcare establishment has advised that the public wash their hands regularly and employ alcohol-based cleaners to disinfect the surfaces. It has been identified that the viruses are comparatively easier to eliminate by rubbing alcohol with diluted H2O2 [33]. In the present review, we have discussed how scientists keep track of the genetic tweaks to SARS-CoV-2 as it is spreading worldwide.
2. RISK ASSESSMENT OF SARS-CoV-2
A structured risk management protocol explores a pathogen's various attributes as well as how it communicates with humans and nature. In the study region, risk evaluations are often performed. However, they are also often important in community health to determine the risks that a newly emerging illness can pose to the general public. Emerging disease risks must be assessed in the same way as novel chemicals synthesized in the drug-production arena. The framework of the biological assessment process is based on many well-defined parameters, including the pathogen, the person, and the context or climate, which influence the policy ruling mechanisms that encompass healthcare decisions in the community health domain.
2.1. Host Variety and Rising Infections
A pathogen's host selection determines whether it can invade only one kind of server or a variety of hosts. How did a bat virus transform itself into a pathogen capable of infecting humans and causing a pandemic? The answer to this question can be found in a characteristic viral trait known as accelerated viral genome mutation, which comprises a set of viral descendants with modified proteins capable of binding to and accessing host cells of various animal species as well as humans. RNA viruses, such as coronaviruses, have particularly high genome modification rates, which allows accelerated diversification at the expense of unsuitable descendants [34]. Virus migration to different characters is completely powered by random mutations and a hit-or-miss host experience. Strengthening a virus's host range provides additional reservoirs, which is advantageous for its development. Spike protein is a particularly essential adaptive tool since it enables the identification of target proteins in various infected cells, not just among several host species but also among different tissues within the same host (tropism) [35].
2.2. Routes and Speed of Transmission
SARS, MERS, and COVID-19 are communicable diseases that spread via respiratory droplets when an infected individual sneezes or coughs. The droplets of different sizes are dispersed and deported with conceivably immense pressure, which permits them to travel to comparatively extended distances. People-to-people transmission of SARS-CoV-2 seems to happen mainly among individuals who are close together, such as family members and their associates, as well as nursing homes, hospitals, and other environments that promote close contact. Further research on the routes of transmission of SARS-CoV-2 is likely to offer more insights on how to protect ourselves. It is of utmost importance to decipher what sparked the current outbreak. Some of the major outbreaks are outlined in Table 1. SARS-CoV in 2002–2003, H1N1 influenza in 2009, and MERS-CoV in 2012 with regional spread and mortality have previously been reported [36-42].
SARS-CoV-2 is a single-stranded, enveloped RNA virus that bears club-shaped glycoprotein projections [43, 44]. Based on its phylogenetic relationship and genomic composition, it is categorized as alpha, beta, gamma, and delta coronaviruses. Genomic studies have revealed that the genetic material of α-CoVs and β-CoVs are possibly derived from bats and rodents [45]. Avian species appear to constitute the genetic origin of δ-CoVs and γ-CoVs. Generally, human coronaviruses are 229E (α-coronavirus), NL63 (α-coronavirus), OC43 (β-coronavirus), and HKU1 (beta coronavirus), while 2019-nCoV, SARS-CoV, and MERS-CoV usually infect animals; however, their transmission to humans has resulted in their evolution into human coronaviruses [46]. Interaction between SARS-CoV-2 spike protein and angiotensin-converting enzyme 2 (ACE-2) of the host exhibits a pattern similar to that of SARS in 2003 [47]. SARS-CoV and MERS-CoV cause severe respiratory syndromes in humans, whereas HCoV-NL63, HCoV-229E, HCoV-OC43, and HKU1 commonly infect humans and cause mild respiratory problems. Recently, studies on the interaction between ACE2 and SARS-CoV have opened new avenues for the development of novel treatment approaches against COVID-19 [48]. Clinical investigations have disclosed that ACE2 is localized in certain human organs (nasal and oral mucosa, lung, stomach, small intestine, colon, lymph nodes, skin, spleen, brain, liver, and kidney). Moreover, investigators have detected SARS-CoV-2 RNA in lung wash, sputum, nasal swabs, throat swabs, and blood [49]. The rapid spread of COVID-19 is attributed to its respiratory and extra-respiratory routes of entry into the human body [50]. The testing of samples from multiple sites could lower false-negative results. More research on SARS-CoV-2 routes of transmission will guide us in developing additional protective measures against the pathogen.
3. EFFECT OF SARS-CoV-2 AND ITS MUTATIONS ON GLOBAL TRAVEL
Mutations are probably completely inconsequential. SARS-CoV-2 emerged as a result of mutations of its ancestors, such as SARS-CoV and MERS-CoV. SARS-CoV-2 is an RNA virus with glycoprotein projections [43, 51]. Recently published studies have suggested that SARS-CoV-2 has mutated into S and L types and many more, with minor changes in the genetic footprint [52, 53]. Understanding the mutation trend allows researchers to reframe the roadmap for developing appropriate diagnostic tests and vaccines.
| Sl. No. | Disease | Year | Pathogen | Individuals Infected Worldwide | Worldwide Mortality | Mortality Rate | Ref. |
|---|---|---|---|---|---|---|---|
| 01 | Influenza (Spanish flu) |
1918-1920 | Influenza A (H1N1) | 500 million | 17.4 million | 0.234% | [92, 93] |
| 02 | HIV/AIDS | 1981-ongoing | HIV | 77.5 million | 34.7 million | 44.77% | [94] |
| 03 | SARS | 2002-2003 | SARS-CoV-1 | 8098 | 774 | 9.55% | [95] |
| 04 | Swine Flu | 2009-2010 | Influenza A (H1N1) | 491,382 | 18, 449 | 1%-4% | [96] |
| 05 | MERS | 2012–2020 | MERS-CoV | 2519 | 854 | 35% | [97] |
| 06 | Ebola | 2014-2016 | Ebola virus | 28, 652 | 13 562 | 47.48% | [98, 99] |
| 07 | COVID-19 | 2019-ongoing | SARS-CoV-2 | 184.27 million | 3.97 Million (July, 2021) | 2.47% | [100] |
COVID-19 outbreak was first reported in Wuhan, China. With the passage of time, scientists have identified the S and L strains of SARS-CoV-2. L strain is aggressive, while the S strain is the less prevalent version. Genetic footprint studies suggest the presence of a common ancestor for all these viruses, with limited genetic variation [54]. Multiple introductions may lead to more genomic diversity. Since it is an RNA virus and DNA is its basic genomic setup. By mingling with the host machinery and mismatch coding, great genomic diversity is expected.
Complete genomic sequencing of SARS-CoV-2 from the samples of infected patients in different countries has shown immense genetic variation compared with the strain initially detected in Wuhan, China. Pathological studies performed on three genomic sequences of SARS-CoV-2 isolated from the first patients to be diagnosed in Valencia have shown that the virus has had sufficient time after arriving in Europe to mutate and become distinguishable from the original strain detected in China in January 2020 [55-59]. Virologists across the globe are sharing data and analytical results to arrive at a conclusion and may design a future roadmap.
Moreover, one of the new mutations (N501Y) has been reported to be located within the receptor binding area, and this variant belongs to the Nextstrain clade 20B [60, 61], GISAID clade GR [62, 63], and lineage B.
3.1. Genomic Properties of the New SARS-CoV-2 Variant
A new variant of SARS-CoV-2, SARS-CoV-2 VUI 202012/01, has been reported in the UK. This variant is characterized by distinctive spike protein mutations (deletion 69-70, deletion 144, N501Y, A570D, D614G, D1118H, P681H, T716I, and S982A) that are present in addition to the mutations in other genomic sections [64]. The non-synonymous mutations in the spike protein are more advanced than the random mutations [65, 66].
3.2. Possible Sources of SARS-CoV-2 Variants
A sustained SARS-CoV-2 infection in a single patient, possibly with diminished immuno-competence, is one possible reason for the appearance of the mutation, which is close to what has previously been reported [67, 68]. As a result of the sustained infection, immune-escape mutants tend to accumulate at a rapid pace. The variability in the development of an infection that occurs in a variety of vulnerable species of animals and is then spread back to humans by the host species is another possible rationalization. As a result of transmission within minks in Denmark, a variant with numerous spike protein mutations (which include RBD mutation Y453F and deletion 69-70) emerged [69]. In the Netherlands, too, several spike protein mutations associated with mink have been identified [70]. South Africa has recorded a dramatic rise in a variant with the spike protein modification N501Y, two extra RBD mutations, and several unspecified spike protein mutations after October 2020, as documented in the GISAID EpiCoV [62] database as well as a public press release [71, 72]. A similar rapid increase has been noted since October 2020 of a variant with the spike protein mutation N501Y, two additional RBD mutations, and multiple additional spike protein mutations.
3.3. Possible Impact of the Rate of Variation in Viruses on the Incidence of Re-infections
The latest modifications appear to be linked to the attachment site of the receptors as well as other surface structures that may modify the antigenic features of the virus. It has been suggested that a drop in antibody neutralization will occur depending on the number and position of mutations in the spike protein, but there is little indication that this drop will lead to an elevated risk of re-infection or decreased vaccine efficacy. For a wide variety of mutants with uncertain clinical effects, a certain reduction in neutralization has been found to date with the use of convalescent sera and monoclonal antibodies [67, 68, 73, 74]. It is not known whether this reduced neutralization is linked to the elevated prevalence of re-infection with VUI 202012/01 and whether it is likely to have an effect on the ongoing vaccination drive.
3.4. Possibility of a Wider Spread of the Latest Virus Variant Across the World
Any additional transmissibility will raise the risk of dissemination, especially when the mingling of extended families and the public that is common in this period of the year is not minimized. The variant could be distributed beyond the United Kingdom, particularly if non-essential movement is not diminished or prevented entirely, and may replace the currently circulating variants in the world.
3.5. Recent Mutations in SARS-CoV-2 and the Emergence of Variants of Concern
Virions evolve constantly, resulting in new types and variants. The majority of such changes are minor and some may even make the virus less deadly; however, a few could make it more infectious and difficult to control by vaccination, such as the delta variant designated by WHO [75]. B.1.617 was designated as a variant of concern by WHO in May 2020, indicating that it spreads more quickly, produces more severe illness, or reduces the efficacy of vaccinations and therapies.
3.5.1. Current Status in India and Worldwide
The COVID-19 epidemic in India began in early March 2020 and is still ongoing. COVID-19 had affected <0.7% of the Indian population prior to March 2021. However, in barely two months, the second wave infected an additional 0.36% of the country’s population. The rapid increase in COVID-19 patients in India is linked to a high incidence of extra variations, which led to clinical trial rejections and antibody evasion [76]. Three important changes in the sequence encoding the viral spike proteins co-occur in variant B.1.617.1, i.e., L452R, E484Q, and P681R. Being a component of the California variants B.1.427 and B.1.429, L452R created an issue in the United States since it protected against the neutralizing monoclonal antibodies X593 and P2B-2F6 [77]. In India, the verified daily additional cases increased from 53 per million population (up to March 2021) to >200 per million population (after March 2021) [78].
According to GISAID data, the B.1.617 variant was first discovered in Maharashtra in October 2020. It was originally considered to be a “double mutant” since it had two mutations, E484Q and L452R. However, researchers now think that the variant exhibits three more genetic changes, i.e., E154K, P681R, and Q1071H. The newest materials were genomically sequenced by the Indian SARS-CoV-2 Consortium on Genomics (INSACOG), a group of 10 laboratories functioning within India's Ministry of Health. Genomic sequencing is a method of mapping an organism’s whole genetic information, in this instance, the genetic code of the virus. After a whopping 11.7 million illnesses and 160,000 deaths, researchers are still looking for alterations. Viruses evolve constantly, resulting in millions of new variants [79].
A latest research in the United Kingdom has discovered that 2 weeks after the second dose, the Pfizer/BioNTech COVID-19 vaccines were 88% efficacious against the variant. The B.1.617 variant was successfully countered after two doses of the AstraZeneca vaccine. The relatively poor protection provided by a single vaccination dosage against the B.1.617.2 strain is a cause for worry, which is why second doses have been introduced. Scientists believe that it is essential to maintain the vaccination even when the viral propagation is minimal to prevent the development of new variants under vaccines. The greater the viral frequencies in the populace, the greater is the probability of a vaccine-evasive variant developing. Hence, in certain regions, the rapid pace of vaccination might dampen viral mutation [80]. On the contrary, the slowness in the pace of vaccination in other countries may accelerate the mutation of the virus.
Viruses undergo genetic modifications to adapt themselves to altered host conditions and/or escape adaptive immune system. Since June 2020, SARS-CoV-2 variants with a D614G mutation have prevailed and are spreading worldwide. The United Kingdom detected a novel mutant strain of SARS-CoV-2 called VUI-202012/01 on December 23, 2020 [81-83]. The viruses appear to be constantly mutating, and their behavior and characteristics have been altered in response to human and environmental interactions (Table 2).
| Sl. No. |
Mutant (WHO Label) |
Pango lineage | Country | Date of designation | Worldwide infection |
|---|---|---|---|---|---|
| 01 | Alpha | B.1.1.7 | United Kingdom | 18 December 2020 | 974,911 |
| 02 | Beta | B.1.351 | South Africa | 18 December 2020 | 27,081 |
| 03 | Gamma | P.1 | Brazil | 11 January 2021 | 47,225 |
| 04 | Delta | B.1.617.2 | India | 11 May 2021 | 126,272 |
3.5.2. Variant of Concern
There is an aftereffect of far and wide sickness transmission, more serious illness (e.g., expanded hospitalizations or passings), and significant abatement in balance by antibodies created all through earlier intense contamination, cutoff of treatments or immunizations, or indicative detecting disappointments for this variation. WHO has been naming the SARS-CoV-2 variants of interest and concern since May 31, 2021, and these are being used with scientific designations in public communications regarding the variants [83]. One or more Pango lineages specify a variant designation as well as any other distinctive spike protein modifications. If the variation is difficult to describe, an alternative description may be given (Table 3).
| Sl. No. |
Mutant (WHO Label) |
Spike Protein Substitutions | Characteristics |
|---|---|---|---|
| 01 | Alpha (B.1.1.7) | 69del, 70del, 144del, (E484K*), (S494P*), N501Y, A570D, D614G, P681H, T716I, S982A, D1118H (K1191N*) | a) ~50% increased transmission [84] b) Hospitalizations and case fatality rates suggest a potential for increasing severity [85]. |
| 02 | Beta (B.1.351) | D80A, D215G, 241del, 242del, 243del, K417N, E484K, N501Y, D614G, A701V | a) ~50% increased transmission [86] b) Slightly decreased sensitivity to monoclonal antibody therapy with bamlanivimab and etesevimab [87] |
| 03 | Gamma (P.1) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I | a) Bamlanivimab and etesevimab monoclonal antibody therapy significantly decreased the susceptibility [87] although alternative EUA monoclonal antibody therapies are available [88]. b) Serum neutralization by convalescent and post-vaccination patients was reduced [89]. |
| 04 | Delta (B.1.617.2) | T19R, (G142D*), 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N | a) Increased transmissibility [90] b) Potential for post-vaccination sera to reduce neutralization [91] |
4. FUTURE PERSPECTIVE AND RISK ASSESSMENT OF COVID-19
A reserved risk evaluation process assesses several aspects of a known pathogen and how it engages with the individual and the surroundings. Risk evaluations are usually done in study environments; however, they are also significant in community health to evaluate the risk that a recently rising infection may pose to the population. The risks associated with emerging diseases must be appraised in a manner similar to that of chemicals synthesized as part of drug advancement. The principle of biological risk evaluation employs distinct standards as well as pathogen, individual, and background or surroundings that influence the strategy resolution building processes, together with healthcare policies in the community wellbeing dominion [84-91].
CONCLUSION
Although the rationales described herein are not entirely established, the NIH scientists hypothesize that the public amid the new coronavirus might be peeling it yet if they do not have indications or if it takes fewer of the virus for an individual to get infected, a metric known as the “infectious dose.” The scientists are at present researching how long the virus survives in boogers, drool, and fecal material and below what humidity levels and temperatures it could sustain itself. Genome analysis of SARS-CoV-2 has disclosed that the virus is undergoing mutations with time and travel.
Globalization has connected the world, but the pool of infected patients in a country is a great challenge. The patients have travel history to different regions; hence, treatment approaches may work differently in different individuals. Vaccine trials might get complicated due to a varied pool of patients with different travel histories and mutated strains of the virus with which they are infected. Mutations have therefore complicated the treatment.
In future, there is a dire need to conduct comprehensive investigational studies on the coronavirus family, collect genomic data of all mutated strains, and examine the clinical symptoms of the patients and the outcomes of various treatment approaches. Our experience with COVID-19 has shown why we need vaccines before and not after an outbreak. Scientists need to intensify their efforts for vaccine development to combat COVID-19. More scientific investigations are to be conducted to understand how COVID-19 is likely to mutate in the future [92-101].
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENTS
The authors thank the Principal, CT Institute of Pharmaceutical Sciences, Jalandhar, and the Administration, CT Group of Institutions, Jalandhar for their continual motivation and encouragement. The authors would also like to thank Dr. H.G. Shivakumar, Mr. Gaurav Thakur, Dr. Riyaz Ali Osmani, Dr. Gangadharappa H V, and Dr. Zulfkar Latief Qadrie for their support in this study.


