All published articles of this journal are available on ScienceDirect.
Identifying Host Cell Gene Expression Modulation as Potential Markers for SARS-CoV-2 Infection
Abstract
Background:
The SARS-CoV-2 emergence in 2019 has caused health, safety, and socioeconomic issues worldwide. Current testing prioritizes viral RNA detection, requiring specialized techniques, training, and time periods, resulting in significant testing limitations. Viral infection can cause changes in host cell gene expression, which vary from virus to virus. Recent research has suggested that SARS-CoV-2-induced gene expression modulations in infected human cells may be differentiated from expressions elicited by other acute respiratory illnesses. Data in this study highlight specific genes that are differentially expressed during SARS-CoV-2 infection. This novel application of individual sample analysis, in connection with global databases, provides robust data for genes that are specifically modulated during SARS-CoV-2 infection. This expression profile would be valuable for SARS-CoV-2 testing, prevention, treatment, and basic virology research.
Methods:
Previously collected COVID-19 surveillance-testing samples from cadets at the United States Air Force Academy were used to quantify the expression of 19 target genes using direct primer-mediated qRTPCR. Additionally, samples were analyzed with RNA-seq to assess the different transcriptomes between uninfected and SARS-CoV-2-infected samples. Results were compared with national databases to confirm the agreement between findings.
Results:
A total of 19 genes were identified to be altered during SARS-CoV-2 infection using in-lab experimental results. This expression profile matched previous research and might uniquely describe SARS-CoV-2 infection. The genes expected to be upregulated according to previous research, IL1B, IFI44L, ACE2, and DUX3, were all upregulated. RNA-seq data confirmed these results and identified 122 other genes significantly different between uninfected and SARS-CoV-2 infected samples. The results have a 93% agreement rate with national databases.
Conclusion:
Despite the availability of vaccines for SARS-CoV-2, the continual mutation and evolution of the virus, the emergence of novel and increasingly infectious strains, and the anti-vaccine sentiment increase the need for safe and rapid testing alternative options. The expression profile of altered host genes during SARS-CoV-2 infection could be extremely advantageous to detect and prevent infection, as well as further research efforts for treatment and understanding of SARS-CoV-2 infection.
1. INTRODUCTION
The SARS-CoV-2 emergence in late 2019 and subsequent global quarantine caused serious health, safety, and socioeconomic issues and lowered quality of life worldwide [1, 2]. Current, accurate PCR testing focuses on the detection of viral mRNA, which is dependent upon specialized techniques, equipment, training, and an optimal window of time for detecting viral mRNA [3, 4]. While reliable, this technique often results in significant limitations to widespread testing. Specifically, for current testing, detectable levels of viral mRNA load usually accumulate 72 hours after exposure, delaying results and causing issues with false negatives [5, 6]. Additionally, PCR testing is subject to primers that match the viral genome. With an RNA virus, such as SARS-CoV-2, mutations occur quickly, and the primers may be less effective with mutations and future variants [7]. Viruses are obligate intracellular pathogens that often modulate and change the host cell in many ways, including the transcriptome [8]. Examining host cell gene expression changes may provide early targets to detect signs of infection before viral mRNA is at high enough loads for detection, offering new techniques for earlier testing as well as bolstering research efforts to understand how viruses alter the host cell gene expression [9].
Viral infection can cause a change in host cell gene expression that is unique for different viruses [10, 11]. Recent data by Mick et al. reported that SARS-CoV-2-induced gene expression modifications of infected human cells may be differentiated from the gene expressions elicited by other acute respiratory illnesses (ARIs) [12]. Understanding the profile of altered genes during active SARS-CoV-2 infection can provide novel insights into how the virus infects the cell and how the cell responds to the infection. Genes that are differentially expressed between uninfected and infected cells may highlight pathways that are either being manipulated by the virus or may be important processes the host cell uses to counter the infection.
SARS-CoV-2 transmission can occur prior to the onset of symptoms or in individuals that are completely asymptomatic [13]. A recent analysis showed that up to 40% of individuals from a confirmed positive database were asymptomatic [14]. Without the detection of symptoms, advanced and early diagnostics would be key to helping identify infected individuals and could result in more successful quarantine and isolation measures, limiting the further spread of the virus. Testing with an increased number of targets and indicators of infection will more rapidly and accurately be able to identify sick individuals and isolate them from the healthy population. Additionally, host cell gene expression should be more stable for detection compared to the constantly mutating and changing viral targets. With a better view of the differences in the human gene expression of SARS-CoV-2 infected cells, a potentially unique gene expression profile could be identified for the novel virus and used for diagnostics and also help understand the virus life cycle. The results from this paper represent novel findings that highlight 19 specific gene targets that are modulated during infection and over 120 other genes that seem to be affected by SARS-CoV-2 infection.
2. METHODS
We used previously collected COVID-19 surveillance-testing samples from cadets at the United States Air Force Academy, consisting of both pre-determined positive or pre-determined negative mRNA samples. All personal identifying information was removed from the samples. All tubes were labeled only with the sample and date of collection. RNA was extracted from samples with Qiagen RNeasy kits following the manufacturer’s protocol. Gene target-specific primers were designed, ordered from IDT, and used in quantitative reverse transcription PCR (qRTPCR). SYBR green qPCR was conducted with the Brilliant III Ultra-Fast SYBR Green qRT-PCR Master Mix (Agilent). A BIORAD CFX 96 Touch thermocycler was used for amplification and quantification. Relative changes in gene expression were measured by calculating the delta Ct values (∆∆CT value) by comparing the target gene Ct value with the human actin (ActB) reference gene in uninfected and infected samples. Table 1 shows all of the primers and their sequences. This method is not new, but the gene targets and the sample origins represent novel findings and results.
| Gene | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| 3 - Gene Model | - | - |
| IL1R2 | GGCTATTACCGCTGTGTCCTGA | GAGAAGCTGATATGGTCTTGAGG |
| IL1B | CCACAGACCTTCCAGGAGAATG | GTGCAGTTCAGTGATCGTACAGG |
| IFI6 | TGATGAGCTGGTCTGCGATCCT | GTAGCCCATCAGGGCACCAATA |
| 10 - Gene Model (Includes above 3) ^ | - | - |
| PCSK5 | TGTGGAGAGCACAGACCGACAA | ACAACGACGTGCTCCAGGTAGT |
| WDR74 | GAAAACAGGCGGCGAACTTCAC | TGCTGAAGTGCTTCACCGTCCT |
| FAM83A | ATCCAGCGCCACTGTGTACTTC | CCGTGAACACATCCATCAGGATG |
| ADM | GACATGAAGGGTGCCTCTCGAA | CCTGGAAGTTGTTCATGCTCTGG |
| IFI27 | CGTCCTCCATAGCAGCCAAGAT | ACCCAATGGAGCCCAGGATGAA |
| KRT13 | GATGCTGAGGAATGGTTCCACG | AGCTCCGTGATCTCTGTCTTGC |
| DCUN1D3 | GGAAGTTCCAGGCTGCAACCAT | CGTGCACAGATTCCGTCAATGC |
| Personally Researched Genes | - | - |
| ACE2 | TCCATTGGTCTTCTGTCACCCG | AGACCATCCACCTCCACTTCTC |
| C9orf117 A.K.A CFAP157 (lung cell) | CAGCAGGAACTGGCTAATGAGC | ACGTCACTGTCCTCTTCATCGC |
| KLK1 (rhinovirus expression) | GACACCTGGAAGGTGGCAAAGA | CATAAGACAGCACTCTGACGGC |
| MUC2 (lung, rhinovirus) | ACTCTCCACACCCAGCATCATC | GTGTCTCCGTATGTGCCGTTGT |
| DUX3 (coronavirus) | CTGCTTTGAGCGGAACCTGTAC | CTGCCTCAACTGGCATGATCTC |
| Zscan4 (coronavirus) | GATGACAGCATAAATCCACCTGC | TTGCTTCTCTTGTGGTTTGGGCA |
| RSAD2 (Influenza, rhinovirus, panviral) | CCAGTGCAACTACAAATGCGGC | CGGTCTTGAAGAAATGGCTCTCC |
| IFI44L (Influenza, rhinovirus, panviral) | TGCACTGAGGCAGATGCTGCG | TCATTGCGGCACACCAGTACAG |
| SERPING1 (Influenza) | GCATCAAAGTGACGACCAGCCA | GTCTCTGTCAGTTCCAGCACTG |
| ActB (reference gene) | CCCTGGACTTCGAGCAAGAG | ACTCCATGCCCAGGAAGGAA |
In order to create a gene expression profile for SARS-CoV-2 infection, qRT-PCR was performed to quantify the expression of 19 specific target genes. The study by Mick et al. suggested that the first three genes would be upregulated during infection and suggested the 10-gene model as a panel of candidate genes that would help correctly identify SARS-CoV-2 infection. Further research on common known viral gene expression resulted in the selection of nine further genes of interest that would serve to distinguish this expression profile from those of the other common ARIs [15-19]. We tested for the expression of each gene in a COVID-negative sample as a control and a SARS-CoV2-positive sample. Each gene test was performed a minimum of three times and compared to a housekeeping gene, human Actin (ActB), to establish reliable fold change (or ∆∆CT value) baseline results.
In addition to qRTPCR testing and calculating the ∆∆CT value, the samples were sent to Genewiz for RNASeq analysis [20]. Three SARS-CoV-2 positive individual samples and three SARS-CoV-2 negative individual samples were analyzed and compared to identify significantly differential expression. Data from the RNA-seq analysis was then compared to national online databases of COVID expression data to identify genes that are uniformly upregulated and those that are downregulated during active infection [21, 22]. The function of each gene was identified using online databases and tools, such as Genecards and BioGRID.
3. RESULTS
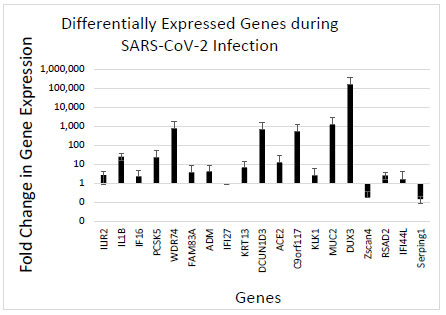
This data represents samples collected during routine surveillance testing. Individual samples were collected from uninfected and infected individuals and were analyzed for gene expression variances in the host cell transcriptome. The data represent a novel healthy population of students from a military academic institution. All of the samples were collected during mandatory surveillance screening, and the individuals did not have symptoms at the time of testing, resulting in a critical dataset that included SARS-CoV-2 negative and asymptomatic SARS-CoV-2, positive individuals. This highlights a major advantage of the dataset and results. A majority of other RNAseq data and results come from SARS-CoV-2-positive and symptomatic patients. The data presented here are exclusively from asymptomatic individuals that were identified as positive from routine testing protocols at an institution of higher education. Samples were screened for 19 different gene target primers that are associated with SARS-CoV-2 infection [12]. Comparing uninfected and infected samples with an internal reference gene resulted in a consistent profile of altered gene expression during SARS-CoV-2 infection (Fig. 1). The data indicate that SARS-CoV-2 infection reliably alters specific genes that may be used as diagnostic markers to confirm infection.
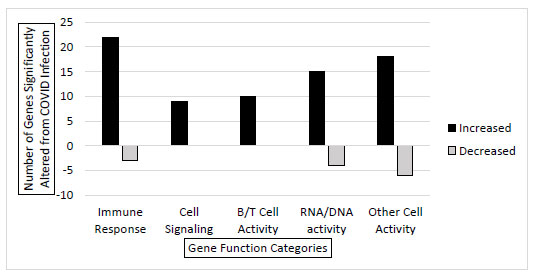
RNA sequencing was conducted on 3 SARS-CoV-2 positive (asymptomatic) and 3 SARS-CoV-2 negative individual samples to get a better comprehensive view of how gene expression is altered in the infected host cell transcriptome. The samples returned with total mapped reads from 3 million to 24 million. The analysis showed that 122 genes had significantly different gene expressions between the infected and the uninfected samples. Evaluating these 122 altered genes, they were put into categories based on gene function. There were significant increases in genes that belong to immune response, cell signaling, B and T cell activity, and RNA/DNA activity functions during infection. Additionally, there were a few genes with decreased expression in immune response and RNA/DNA activity functions (Fig. 2). Collectively, this confirms that SARS-CoV-2 alters the host cell gene expression in significant and consistent patterns during infection.
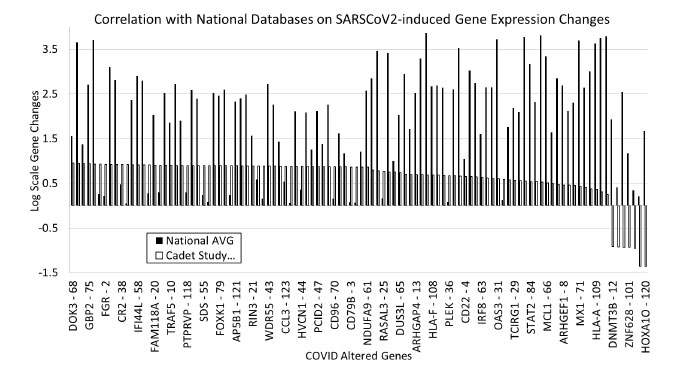
To confirm the consistency of the altered genes in other samples, national databases of COVID gene expression data were screened to identify how the data from our samples compared with national databases. Overall, our cadet data correctly matched the national average trends. There were differences in the scale and amount of gene expression alterations, but 93% of our genes that showed an upregulation within our samples also had increased expression in the national databases. Only 6% of our data had opposite results compared to the national average (Fig. 3).
4. DISCUSSION
The altered gene expression during SARS-CoV-2 infection provides a novel insight into SARS-CoV-2-induced host cell gene expression changes. This profile of modulated genes may serve as a unique diagnostic tool for SARS-CoV-2 infection. The genes identified by Mick et al. in the three gene model, specifically IL1B, IFI44L, and ACE2, were all upregulated in our data, confirming that those three genes are consistently upregulated during active SARS-CoV-2 infection, even with asymptomatic individuals, which is the case in our samples (Fig. 1). Also largely upregulated were DUX3, MUC2, C9orf117, DCUN1D3, and WDR74, all of which correspond to different system regulatory processes, such as lung function or lung cell protein synthesis, which is expected for an ARI. The exact level at which these genes are upregulated may serve to form the unique expression profile for this virus, distinguishing it from other common ARIs. Future work could look at other databases and results to confirm the extent these target genes are modified and if that change is constant across individuals.
In addition to the 19 genes screened through primer-mediated direct qRTPCR (Fig. 1), there were 122 differentially expressed genes as determined through RNA-seq. These 122 genes were categorized based on gene function, and it is not surprising that most of the significantly upregulated genes belong to functional families that involve the immune system, B and T cell function, cell signaling, and DNA/RNA activities. These are all critical functions that the cell would need to activate in order to fight off the viral infection. Individual gene analysis could be conducted to identify the exact role and mechanism in the host cell response or viral manipulation that occurs during the viral infection. Gene expression could vary from individual to individual. Our sample set consisted of 3 infected (asymptomatic) and 3 uninfected young college-age individuals. The data could be further validated with more testing and a wide variety of other individuals. Regardless of the small sample size, the results were validated by comparing other data points in databases.
Gene expression modulation was verified with national databases and had a 93% agreement with our cadet data and national data. Only 6% of genes did not correlate with national averages, and those genes were DNMT3B, FABP6, DYNC1LI1, ZNF628, SERPINB3, MADCAM1, and HOXA1O. These genes are involved with specific metabolic or regulatory roles in the cell and may be an artifact from the individual samples used in the study. Further work needs to be conducted to investigate the role of these genes during viral infection with SARS-CoV-2.
5. CONCLUSION
As this virus continues to disrupt normal societal proceedings and take lives on the magnitude of millions across the globe, the need for safe and fast detection options is paramount. Despite the widespread use of the Pfizer and Moderna mRNA vaccines, factors, such as lengthy delayed distribution and supply chain issues, the evolution of untested and increasingly infectious viral strains, and rising anti-vaccine sentiment continue to increase the need for safe and rapid testing options, especially as people become less fearful of the consequences of infection. An expression profile that can easily differentiate SARS-CoV-2 from other ARIs and determine viral infection status before the time period for the presence of detectable levels of viral mRNA could be extremely advantageous to quarantine, isolation, and prevention efforts, as well as continued research efforts for the treatment and vaccination against SARS-CoV-2. This study serves as a proof of concept and starting point for advances in using host cell modulation of gene expression as a novel way to detect infections.
LIST OF ABBREVIATIONS
| ARIs | = Acute respiratory illnesses |
| qRTPCR | = Quantitative reverse transcription PCR |
| ActB | = Human Actin |
| MUC2 | = lung, rhinovirus |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study is in accordance with the Institutional Review Board Statement: Determined to be Not Human Subject Research: FAC20210016N. Also, this manuscript has been approved for public release with PA number: USAFA-DF-2022-379.
HUMAN AND ANIMAL RIGHTS
No humans or animals were used that are the basis of this study.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Data and materials are available by contacting the corresponding author.
FUNDING
This study is financially supported by the USAFA Department of Biology, Defense Health Agency Grant to the Life Science Research Center at USAFA.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
The authors would like to thank the Biology department faculty, staff, and students for supporting this work. They would specifically like to thank Cadet Antonio Cruz, Cadet Cosmo Cao, and Francesca Iova for their assistance with this project. Additionally, Col. Steven Hasstedt and Lt. Col. David Morris provided critical support and resources to help this project.
The views expressed in this paper are those of the authors and do not necessarily represent the official position or policy of the U.S. Government, the Department of Defense, or the Department of the Air Force.


