All published articles of this journal are available on ScienceDirect.
Omicron Plus India: Trends Observed in S-gene Target Failure Detection Rate
Abstract
Background:
COVID-19 is an infectious disease caused by SARS-COV-2. As the world eagerly awaits the transition of the COVID-19 pandemic to an endemic, the ever-evolving virus has a new variant of concern, as announced by WHO on November 26th, 2021. The Omicron variant, B.1.1.529, was first reported to WHO on November 24th, 2021. The variant has a large number of mutations in the S-gene and has caused a detrimental change in COVID-19 epidemiology. The efficient management of COVID-19 depends on early diagnosis and treatment. This new chain of new variants of concern has always posed a challenge to the accurate diagnosis of COVID-19.
Methods:
In this study, we studied the S-gene target failure of positive cases of COVID-19 from December 2nd, 2021, to January 1st, 2022. Moreover, we checked this trend per month for two months after that by accumulating positives. All positive samples detected for COVID-19 by ICMR approved kit (target selections N and ORF1ab) were re-run using another ICMR-approved kit with three targets (S, N, and ORF1ab). The absence of S-gene in the target, i.e., S-gene target failure (SGTF), can be considered an indication of an Omicron variant as per the advisory policy. In continuation with this, considering the next wave in China, we tested SGTF for the next two months, and as expected, a reduction in SGTF percentage was observed.
Results:
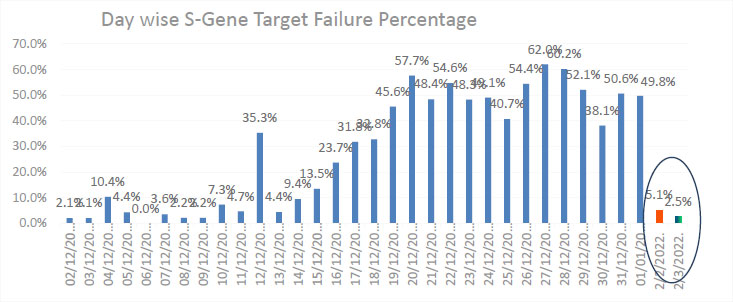
A total of 53,276 samples were received in December 2021, of which 4848 samples showed a positive result for SARS-CoV-2. Out of this, 2119 (43.70%) samples showed SGTF. To analyze this trend, we tested 156 samples from February, 2022 and 118 samples from March, 2022, which resulted in only 8 (5.1%) and 3(2.5%) samples showing SGTF.
Conclusion:
The study sheds light on the doubling rate of infection of COVID-19 in India and SGTF, indicating the spread of the Omicron variant. The infection can be controlled by accurately diagnosing variants of concern (VOC), but all positive samples cannot be screened for sequencing. In this case, kits with multiple targets that show the SGTF can be used as a proxy screening test, and for further confirmation, whole genome sequencing can be done.
1. INTRODUCTION
COVID-19 has caused more than 286 million infections and 5.4 million deaths to date. COVID variants have appeared at regular intervals during this pandemic, such as alpha, beta, gamma, delta, and Omicron [1]. China has recently extended COVID-19 restrictions in its largest city, Shanghai, in the second week of April 2022, after new COVID cases surpassed 13,000 for the first time. The two-stage lockdown, which was originally set to end in Shanghai's western districts, has now been extended until further notice. India waived all COVID-19 restrictions on March 31st, 2022. One should not forget that other countries like China, the United States, and other South Asian countries have seen a massive rise in COVID cases. In the past, while these countries experienced a peak in COVID cases, India was COVID-free, and when these countries started observing a decline in COVID cases, it was India that began witnessing a rise on the other hand.
A new variant of COVID-19 keeps appearing, but still, the world is looking forward to an end to the COVID pandemic. The latest variant XE, Omicron’s subvariant, is considered more transmissible than other variants; it is the new variant of Omicron that was first detected in the UK. The XE variant is a recombinant of both subvariants BA.1 and BA.2 of Omicron. Until now, India has reported two cases of the XE variant [2].
Omicron has quickly replaced the Delta strain in South Africa, accounting for more than 37,000 cases in a record time of just 20 days, compared to a maximum of 26,000 infections per day in all previous waves [3]. On November 24th, 2021, Omicron variant B.1.1.529 was first reported to WHO, and on November 26th, 2021, WHO classified Omicron variant B.1.1.529 as a variant of concern (VOC). This classification was done based on the primary information from South Africa provided by the Technical Advisory Group on virus evolution, indicating a large number of mutations and detrimental changes in COVID-19 epidemiology [4]. The Omicron variant has more than 30 mutations in the spike protein alone. The WHO has reported that preliminary evidence suggests an increased risk of transmission compared to other VOCs [5, 6].
Of the overall infections, in London, more than 90% of the patients had Omicron. Similarly, the United Kingdom was also severely affected, with more than 100,000 cases reported daily [7]. Even with aggressive lockdowns, restrictions, and a push for booster vaccinations, the same trends were seen in France, Germany, the Netherlands, and other European countries [8]. The trend was not different in the US; the variant swiftly spread to the country and added to the ongoing Delta wave. In the last week of December 2021, more than 4,00,000 COVID-19 cases were recorded daily in the US, with Omicron causing 58% of overall COVID-19 cases [9-19]. Omicron, which spreads rapidly, was recorded in more than 100 countries and caused the global burden to exceed 1 million daily on December 28th, 2021. Omicron has been reported in India, initially in patients who had returned from international travel, but now it has been reported in patients without a travel history as well. There was a sudden surge in infections in Mumbai and Delhi from December, 2021 to January, 2022 [10].
The presence of mutations in the COVID-19 virus in the patient sample can impact the test performance. The impact of mutations on the test performance can also be influenced by several factors, including the prevalence of VOCS in the population, their sequence, and the design of the test [6]. These conclusions and information lead to renewed research to examine the variant’s impact on the efficacy of existing vaccines and tests. The WHO, FDA, CDC, and European Centers for Disease Control reported that using the SGTF of PCR assays as a proxy for the variant helped in identifying Omicron [5, 6]. The Omicron variant has a 69-70del mutation in the S-gene region. This mutation causes a target failure of the S-gene in the results of the kits, which have multiple targets that can indicate to clinicians and scientists a possible Omicron variant. Such samples can be sent on a fast-track basis for confirmation of the variant by sequencing.
2. MATERIALS AND METHODS
2.1. Settings
Samples were collected by Thyrocare Service Providers (TSPs) and transported at the desired temperature with mandated registration at Thyrocare, Navi Mumbai, India. Sample storage, RNA extraction, PCR setup, and data analysis were performed on-site. The duration of the study was from December, 2021 to March, 2022.
2.2. Study Population and Specimen
This is a single-site study; a total of 53,276 samples were screened for the presence of SARS-CoV-2, and 4848 positive samples were included in the study from December, 2021 to January, 2022. To check for the data during February and March, 2022, we selected two days, i.e., 2nd February and 3rd March and found 156 and 118 positive samples, respectively. After routine processing (targeted for the presence of N gene, ORF1ab) and reporting of the samples, all positive samples were advanced to RT PCR (targeted for the presence of N gene, ORF1ab, and S-gene) as per protocol.
2.3. Statistical Analysis
For statistical analysis, guidelines were followed as per the STARD recommendation [11]. Positive agreement and negative agreement were verified for qualitative results. The doubling rate of the SGTF is represented using a bar graph.
2.4. COVID-19 RNA Extraction, PCR Assay Setup, and Result Interpretation
Nucleic acid extraction (200-μL sample input volume) was performed using MagMAX™ Viral/Pathogen Nucleic Acid Isolation kit (Applied Biosystem -Thermo Fisher) by using a semi-automated Thermo Kingfisher Flex extraction system. The PCR assay setup was done using QIAgility, a liquid dispenser automated system by QIAgen, which was used for mixing RNA and already prepared master mix. The PCR assay was done using various commercially available ICMR-approved kits, and subjected RT PCR amplification was done using Quantstudio-5 (Thermo). The results were analyzed using Quantstudio design and analysis software version 1.5.1. All procedures were performed as per the manufacturer’s kit instructions [12-15].
| S.No. | Date | No. of the Samples Positive | No. of Samples with SGTF | % SGTF |
|---|---|---|---|---|
| 1 | 02/12/2021 | 48 | 1 | 2.08 |
| 2 | 03/12/2021 | 48 | 1 | 2.08 |
| 3 | 04/12/2021 | 48 | 5 | 10.42 |
| 4 | 05/12/2021 | 46 | 2 | 4.35 |
| 5 | 06/12/2021 | 34 | 0 | 0.00 |
| 6 | 07/12/2021 | 56 | 2 | 3.57 |
| 7 | 08/12/2021 | 46 | 1 | 2.17 |
| 8 | 09/12/2021 | 46 | 1 | 2.17 |
| 9 | 10/12/2021 | 41 | 3 | 7.32 |
| 10 | 11/12/2021 | 43 | 2 | 4.65 |
| 11 | 12/12/2021 | 17 | 6 | 35.29 |
| 12 | 13/12/2021 | 45 | 2 | 4.44 |
| 13 | 14/12/2021 | 53 | 5 | 9.43 |
| 14 | 15/12/2021 | 52 | 7 | 13.46 |
| 15 | 16/12/2021 | 59 | 14 | 23.73 |
| 16 | 17/12/2021 | 44 | 14 | 31.82 |
| 17 | 18/12/2021 | 61 | 20 | 32.79 |
| 18 | 19/12/2021 | 57 | 26 | 45.61 |
| 19 | 20/12/2021 | 78 | 45 | 57.69 |
| 20 | 21/12/2021 | 93 | 45 | 48.39 |
| 21 | 22/12/2021 | 108 | 59 | 54.63 |
| 22 | 23/12/2021 | 116 | 56 | 48.28 |
| 23 | 24/12/2021 | 110 | 54 | 49.09 |
| 24 | 25/12/2021 | 91 | 37 | 40.66 |
| 25 | 26/12/2021 | 90 | 49 | 54.44 |
| 26 | 27/12/2021 | 234 | 145 | 61.97 |
| 27 | 28/12/2021 | 344 | 207 | 60.17 |
| 28 | 29/12/2021 | 453 | 236 | 52.10 |
| 29 | 30/12/2021 | 609 | 232 | 38.10 |
| 30 | 31/12/2021 | 808 | 409 | 50.62 |
| 31 | 01/01/2022 | 870 | 433 | 49.77 |
| 32 | 02/02/2022 | 156 | 8 | 5.13 |
| 33 | 02/03/2022 | 118 | 3 | 2.54 |
| Total | 4848 | 2130 | ||
3. RESULTS
The selected 4848 positive COVID-19 samples were detected using the targets N and ORF1ab. These samples were re-run using a combo kit with targets of S, N, and ORF1ab. This step was included to check the presence or absence of S-gene target criteria by advisory bodies. Of the total samples processed, 2119 (43.71%) samples showed SGTF (Table 1 and Fig. 1). The infection transmission rate, i.e., the positive rate and the corresponding SGTF, clearly showed the high doubling rate of the Omicron variant in India. We tested 156 and 118 positive samples in February and March, 2022, respectively, and found SGTF in only 8 and 3 samples. The result clearly demonstrated a sudden drop in Omicron cases from 43.7% to 5.1% and 2.5%, respectively.
4. DISCUSSION
India has a nearly 1.3 billion population and is the 22nd most visited nation in the world. Therefore, there is significant interest globally in the possible course and consequences of all COVID waves. India has recently witnessed a devastating second wave due to the Delta variant, which claimed about 0.33 lives besides causing tremendous damage to the socio-economic structure [16].
The data showed an increased rate of infection with COVID-19 in India. The infection transmission was low in the first week of December, but the infection started to spread, and a doubled infection rate was observed in the second week of December (Fig. 1). SGTF indicated the Omicron variant spread, and the data after December 10th reported a very high transmission rate of Omicron, which was similar to many other countries [1]. Omicron is a highly transmissible variant, with studies from South Africa and the UK reporting a doubling in infection rates of 3.38 d (95% CI: 3.18-3.61 d) and 2-2.5 d [1]. The rapid surge in the daily number of infections, compared to global trends, strongly suggested the dominance of Omicron-variant infections in India. However, global Omicron trends suggest a decline in case fatality rates and hospitalizations compared to the Delta variant.
The Omicron variant B.1.1.529 has a large number of mutations in the S-gene and has caused a detrimental change in COVID-19 epidemiology. Our study included the COVID positive cases, and data demonstrated that 43.7% of positive samples possessed SGTF suggestive of Omicron from December, 2021, while it was 5.1% from February, 2022 and 2.1% from March, 2022.
The Omicron variant includes four Pango lineages: the parental B.1.1.529 and the descendant lineages BA.1, BA.2, and BA.3. The BA.1 lineage, which accounts for 97.4% of sequences submitted to GISAID as of January 2019, whereas BA.3 (only a few dozen sequences) has the 69-70 deletion in the spike protein, while BA.2 does not have any. Knowledge of B.1.1.529 is still developing, but this lineage is more diverse, with the 69-70 deletion present in nearly 80% of all currently available sequences. The presence of the 69-70 deletion in the spike protein causes a negative signal for the SGTF in certain PCR assays. This SGTF can be considered a marker suggestive of Omicron. Concerning which Omicron lineages are circulating locally, there is a possibility of missing cases of BA.2 or other isolates lacking the 69-70 deletion. Therefore, confirmation should be obtained by sequencing at least a subset of SGTF samples as this deletion is also observed in other variants (e.g., alpha and subsets of Gamma and Delta), which are circulating at low levels worldwide [17].
Even though India is now not recording significant COVID cases, as per the National Center for Disease Control (NCDC), the prevalence of Omicron sublineage BA.2 is gradually increasing in India. Earlier, the BA.1 variant was dominant among the samples collected from travelers. As per references in community settings, the BA.2 subvariant is gradually increasing [18, 19].
Despite indications of the rapid growth and decline in cases, along with lesser hospitalizations and deaths compared to the second wave, a major concern is the emergence of a new variant due to rapid community transmission, thus changing the projected course of the pandemic. Furthermore, the predicted caseload changes depend on the change in the testing protocol. The projections also need to be updated with the availability of new data. Many variants of the SARS-CoV-2 coronavirus have arisen throughout the pandemic. Some spread worldwide, while others quickly faded away or were supplanted by other variants. Viruses mutate all time, but only some mutations affect their ability to spread, evade prior immunity from vaccination or infection, or the severity of the disease. Therefore, additional mutations need to be further studied to understand their impact on immune escape potential.
CONCLUSION
In conclusion, this study sheds light on the doubling rate and trend of COVID-19 infection in India and the SGTF failures, indicating the spread of the Omicron variant. Control of infection can be achieved through accurate diagnosis of VOC, but all positive samples cannot be screened for sequencing. In this case, the kits with multiple targets showing the SGTF can be used as a proxy screening test and for further confirmation, whole genome sequencing can be performed.
LIST OF ABBREVIATIONS
| SGTF | = S Gene Target Failure |
| VOC | = Variant of Concern |
| NCDC | = National Centre for Disease Control |
AUTHORS' CONTRIBUTIONS
PKG performed testing and wrote the manuscript; KK contributed to copy editing and manuscript verification. CN contributed to selecting the study design and verification of data and manuscript. CS participated in the verification of data and manuscript. PS, DP, and RA performed the study.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was carried out on the samples identified by a laboratory-generated number with no traceability to patients. All patient details were thus kept confidential. Moreover, further analysis for SGTF was carried out on the leftover RNA of the extracted samples after releasing the reports. Thus, the results of this test did not affect the patient’s diagnosis.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data generated and/or analyzed during the current study are not publicly available due to patient confidentiality but are available from the corresponding author on reasonable request [C.N].
FUNDING
The study is funded by Thyrocare Technologies Limited.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the support of the Thyrocare Technologies Limited team and would like to thank their colleagues who participated in this study.


