All published articles of this journal are available on ScienceDirect.
Molecular Mechanisms of Non-Coding RNAs in Modulating the Pathogenesis of SARS-Cov-2 Infection
Abstract
The coronavirus disease 2019 (COVID-19) has been spreading worldwide since December 2019. It is a significant threat to community healthcare in all countries worldwide, so policymakers and researchers are paying close attention to it. The most significant components of non-coding RNAs (ncRNAs) are miRNAs and lncRNAs, which serve as regulatory elements. They are vital ingredients of the transcriptome, with a role in normal biological reactions and inflammatory processes, including viral infection. In the field of viral infection, microRNAs and non-coding RNAs with 19 to 25 nucleotides receive more attention as they target mRNAs to control gene expression. However, the role of many lncRNAs is yet to be discovered. In this review, we provide detailed information about the effects of host lncRNAs and viral lncRNAs, interactions between lncRNAs and their interactions with other ncRNAs, and small membrane vesicles called exosomes and microRNAs in COVID-19 infection. The profile of ncRNAs in host cells of the SARS-CoV-2 virus is altered. As a result, these changes may serve as valuable indicators for disease development and severity. Understanding these pathways will help researchers learn more about SARS-CoV-2 pathogenesis and seek more practical treatments to control cytokine storm and viral life cycle.
1. INTRODUCTION
The coronavirus disease 2019 (COVID-19) outbreak has been the most significant public health epidemic since 1918 and has caused many economic and health issues [1-3]. Therefore, learning more about its features and interactions with human host cells is essential to discover efficient therapies. MERS-CoV and SARS-CoV-1 are two additional dangerous human coronaviruses that cause moderate respiratory tract disease with a 36% and almost 10% mortality rate, respectively [4]. However, the severity of COVID-19 differs among patients. The vast majority experience common cold symptoms that develop into a mild pneumonia case, while about 14% of persistent cases express severe symptoms such as shortness of breath [5]. All strains of single-stranded, positive RNA (+ssRNA) viruses make up most of coronaviruses [6], via an electron microscope, the presence of spike glycoproteins on the coronavirus envelope gives it a crown-like appearance (Corona is the Latin word for crown) [7, 8]. When adapting to their new human hosts, SARS-CoV-2 is susceptible to genetic evolution, with mutation development over time, resulting in variants with different features than their progenitors [2].
Nowadays, non-coding RNAs (ncRNA) are used in genomic medicine [9]. Regulatory elements such as microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) are non-coding RNAs that control gene expression rather than actively engaging in cell physiological activities [10]. lncRNAs are transcripts with a length of 200 nucleotides or more with various roles in cell biology [11]. The identification of lncRNAs as potential major regulators of inflammatory genes suggests that they may be required for regulating inflammatory responses [12-14]. Understanding the effect of their altered expression levels and mechanism of action can have a substantial impact on immunology and infectious diseases. Scientists discovered that 500 lncRNAs interfered with the cell response to viral infection by examining SARS-CoV-infected cells [15]. They also suggest that these non-coding characters in host cells and RNAs have an essential modulating role in the antiviral innate immune system. The majority of lncRNAs associated with viral infections co-express genes involved in maintaining lung homeostasis, and they have been linked to IFN mechanistic pathways [16].
MicroRNAs are non-coding RNAs with 19 to 25 nucleotides that target mRNAs and cause translational repression or mRNA destruction to regulate gene expression. miRNA is a potent gene regulator that affects virtually every aspect of gene regulation [17, 18]. miRNA regulates gene expression by targeting the 3' and 5' untranslated regions (3' UTR and 5' UTR, respectively) (UTR) and coding regions [19-21]. Furthermore, microRNAs significantly impact the production of cytokines, chemokines, and growth factors [22]. Plants, animals, and viruses contain microRNAs with various biological roles [23, 24]. MicroRNA-targeted therapies have been suggested to treat malignancies, viral diseases, and other illnesses since they are essential regulators of gene expression [25]. Many miRNAs have been identified as markers in virus-infected illnesses [26]. MicroRNAs could be encoded by viral genomes, encompassing DNA and RNA viruses. MiRNAs generated from viruses may also be synthesized in host cells with a function in the life cycle of a virus as well as cellular consequences [27, 28]. On the other hand, miRNAs may attach to complementary regions on the viral RNA sequence, thus reinforcing the viral genome's silencing effect and reducing protein production [29].
With the aid of miRNA response elements, lncRNAs and circRNAs competitively attempt to bind miRNAs and form competing for endogenous RNAs (ceRNAs) [30]. CeRNAs may act as miRNA sponges, controlling the production of miRNAs that target specific mRNAs. The discovery of lncRNA/circRNA aids the development of a ceRNA network, potentially contributing to the discovery of new and effective treatment targets [31].
2. INTERPLAY BETWEEN COVID-19 AND NON-CODING RNAS
SARS-CoV-2 has produced a large epidemic with a significant mortality rate throughout the globe, and death rates are continuously increasing [32]. Understanding the relationship and effect of this virus on human host cells is critical [7]. In the coronavirus genome, spike, envelope, membrane, nucleocapsid, and all structural proteins are encoded by positive-sense RNA [32]. The rapid mutation rate in the spike nucleotide sequence has been suggested as one of the causes of SARS-CoV-2’s high transmission rate [33].
Viral resistance is affected by the ncRNA network of host cells. Infection typically alters the expression patterns of ncRNA in the host, potentially increasing viral proliferation and propagation conditions. As a result, ncRNAs boosted during the infection may be efficient biomarkers for disease progress and severity [7]. Furthermore, viruses are capable of aggressive conflict against host cells to deplete their metabolic resources required for viral proliferation. They generate exogenous ncRNAs, dysregulating the expression of hundreds of host genes related to metabolism control. The ncRNAs encoded by a number of these affected genes are an essential part of the virus-induced pathogenic transcriptome in host cells [34].
LncRNA transcripts with poly(A) 3′ ends and 5′-capped terminals [21] seem incapable of encoding proteins. In the human genome, there are 16,193 lncRNAs discovered that could be transcribed from both strands, according to the most current GENCODE V30 release, but only about 3% of annotated lncRNAs have been assigned a function [35, 36]. RNA polymerase II and III are accountable for their transcription and are comparable to mRNAs in terms of evolutionary conservation. However, RNA polymerase II transcribes the bulk of them [30, 37]. LncRNAs have a role in chromosome shaping, miRNA generation, and mRNA suppression [38]. They may interact with proteins, RNA, DNA, or a combination of these molecules to mediate their activities, and their secondary structure and/or sequence might influence the lncRNAs' responsibilities (Fig. 1) [21].
3. ROLE OF LNCRNAS IN SARS-COV-2 INFECTION
LncRNAs that moderate viral proliferation is categorized into two groups according to their origin: host-encoded lncRNAs and virus-encoded lncRNAs [20]. Host-virus lncRNA interactions typically occur in the cell's cytoplasm, nucleus, and extracellular spaces [38].
In the first stages of viral infection, host cells begin their antiviral response by evolving their lncRNA profile, and after infection, these RNAs express differently. They play a role in signaling pathways regulating the cell cycle, programmed cell death, immune response, and gene expression [21]. In addition, viruses may change the endogenous lncRNA expression of host cells. In contrast, there is no indication that SARS-CoV-2 can generate any long non-coding RNA (lncRNA) [15]. So far, most lncRNA research has been focused on cancer. Meanwhile, innate immunity research on lncRNAs has been poor, accounting for just 4% of all lncRNA articles published so far [19].
The innate immune system contributes to the formation and maturity of the adaptive immune system by acting as a relatively quick response to certain infections by inducing inflammation [39, 40]. Early in the infection process, the SARS-CoV-2 enters T lymphocyte cells and destroys them, leaving patients with severe lymphopenia. As a result, more lymphocytes target the heightened inflammatory responses of the innate and adaptive immune systems, which result in their death [41]. The regulation of the innate antiviral immune response of host cells is mediated by lncRNAs [15].
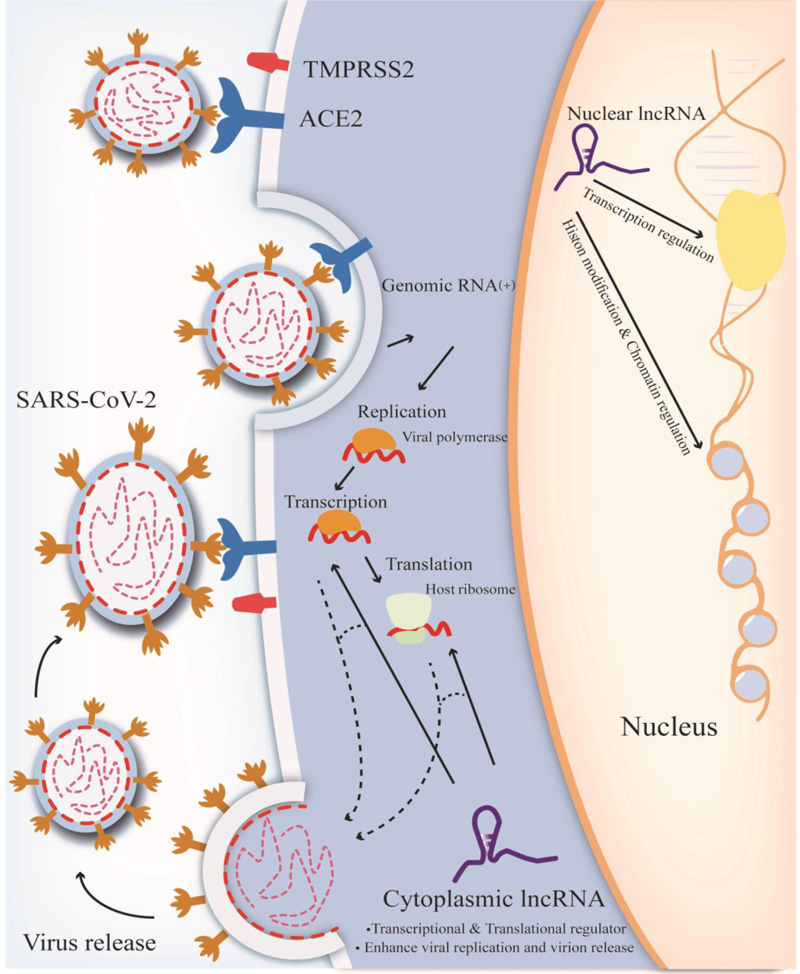
The expression of lncRNAs is related to type I interferon receptor, signal transducer, and activator of transcription 1 in the same way it occurs in influenza-infected cells [15]. JAK/STAT, NF-KB, HIF-1A, and MAPK are pathways that may regulate the synthesis of interleukins through lncRNAs [32, 42, 43]. In infected cells with SARS-CoV-2, host-derived lncRNAs such as MALAT1 and nuclear-enriched autosomal transcript 1 (NEAT1) may also be used as infection biomarkers [44, 45]. Overexpression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA was observed in SARS-CoV-2 infection, which is also activated in various neoplastic disorders and post-lung transplant inflammatory conditions [43]. MALAT1 silencing diminishes the occurrence of SARS-CoV-2 patients' cytokine storms by inhibiting neutrophil chemotaxis [43]. The expression of both MALAT1 and NEAT1, which are immunomodulatory lncRNAs, is altered in patients with severe symptoms. A study suggests that NEAT1, MALAT1, and an antiapoptotic lncRNA named MTRNR2L12 are significantly overexpressed in the bronchoalveolar lavage of severe patients, while their expression is suppressed in patients with mild symptoms. NEAT1 is only found in sites of inflammation and infection [46]. On the one hand, MALAT1 seems significantly expressed in CD4+ T cells of individuals with moderate symptoms, illustrating its protective role in T cells. As a vital protein for the COVID-19 virus to enter the body, cathepsin L (CTSL) may be the potential mechanism for the initiation of these lncRNA-mediated inflammatory responses [47].
Another lncRNA linked to virus-induced inflammation is DANCR, indicating a dramatic drop in inflammation-prone lung tissues [48]. It can also regulate the function of catenin beta-1, which is a protein that is linked to respiratory illnesses [49]. Generally, NEAT1 and DANCR can interact with many ncRNAs, sponge miRNAs, and transcription factors such as STAT3 to inhibit inflammation [50]. Through the IL-11/JAK2 pathway, DANCR can activate STAT3, a pro-inflammatory transcription factor [50]. STAT3 triggers IL-1b, IL-6, NFkB1, and SPI-1, which are also transcription factors interacting with TNF, DANCR, and the NEAT1-associated protein DANCR [51-54]. Downregulation of DANCR in infected bronchial epithelial cells is linked to a reduced ACE2 level [55].
Because lots of pathways involving miRNAs, lncRNAs, and mRNAs have been reported to have essential roles in characterizing cellular activities in the course of a viral infection, it's vital to study these connections in a unified manner to fully understand the regulatory non-coding RNA networks that underpin SARS-CoV infection [56].
4. ROLE OF MICRORNA IN SARS-COV-2 INFECTION
Viruses are dependent on host cell activity for many stages of their life cycle. The miRNA pathway is an essential part of the host's regulatory system [57]. Viruses may interact with the host miRNA in multiple ways [58]: 1) Host miRNAs that bind directly to the viral RNA and may control viral translation or other elements of the viral life cycle. Some of these miRNAs may have an antiviral impact, allowing the immune system to combat the disease or causing the virus to enter a dormant condition. 2) The virus may alter host miRNA expression, influencing host or viral RNA targets. 3) Some virus may encode their miRNAs, which control the RNA targets of the host or virus and may alter the host's signaling pathways, allowing the virus to survive and replicate. Some viruses cause infected cells to mutate, so there are many links between viral infection and miRNA expression [59, 60].
To learn the link between miRNA and COVID-19, we must first comprehend the pathogenic mechanism of SARS-CoV-2. Inflammatory cytokines, including interleukin-6 (IL-6), TNF-α, and inflammatory complexes like the inflammasome, are produced due to ACE-mediated SARS-CoV cell entry [61, 62]. The main contributors to the inflammatory cytokine storm appear to be the inflammasome, IL-6, and NOD-like receptor protein 3 (NLRP3) [63, 64]. miRNA analogs suppress the production of proteins related to the COVID-19-mediated cytokine storm [65]. Fabbri et al. have discovered that miR-93-5p inhibits the IL-8 gene. They discovered that a) the miR-93-5p level is increased in the cells and b) IL-8 mRNA content and IL-8 output were significantly reduced when pre-miRNA sequences were transfected into different cell lines [66]. According to Oglesby et al., interleukin-8 production is reduced when miR-17 is overexpressed in airway epithelial cells [67]. According to Hong et al. research, polyethylenimine (PEI) was used to transport plasmid DNA encoding miR-200c into target cells, resulting in increased production of miR-200c and efficient suppression of IL-6, IL-8, and CCL-5. Hong et al. also discovered that miR-200c targets the 3'UTR of IL-6, IL-8, and CCL-5 [68].
5. MIRNA AND LNCRNA INTERACTION
MiRNAs can regulate lncRNAs expression. By incorrect base-pairing, RNA-induced silencing complexes (RISCs) may be used to attack lncRNAs, diminishing their morphological and chemical stability [69]. On the other hand, through specific pathways, miRNAs enhance their expression and mature cytoplasmic miRNAs reach the nucleus and influence the transcription of mRNA and ncRNAs [70]. For instance, mature miR-140 in stem cells generated from adipose tissue enhance NEAT1 expression and its stability through binding to particular sites on the NEAT1 locus [71].
Furthermore, viral miRNA can alter the host cell microenvironment by post-transcriptionally regulating many host protein-coding transcripts and lncRNAs [38]. In terms of viral infection, the lncRNA H19 can bind to the let-7 miRNA family, lowering the cell's supply of let-7 and rendering it more susceptible to infection [6]. Many forms of cancer cells have high levels of H19, making them vulnerable to viral infection [7]. The host cell transcriptome is activated during viral infection due to the infected cell's natural immunological response [72].
In turn, lncRNAs can function as miRNA sponges and may be utilized as endogenous RNA that competes with miRNA function [56]. They may bind to target miRNAs in ceRNA networks through miRNA reaction components (MREs), preventing miRNA-mediated degradation of targeted mRNAs [73]. They also produce miRNAs by splicing RNA for post-transcriptional control of mRNAs as miRNA precursors [74]. Consequently, by vying with miRNAs for particular targets, identification, and attachment to the 3'UTR of target mRNAs, lncRNAs may impede miRNA-mediated negative regulation of target mRNAs. However, the function of lncRNAs and miRNAs produced by viruses and host cells in viral infection and their competitive binding to mRNAs remains largely unknown, necessitating additional research [20].
6. CIRCRNA AND LNCRNA IN COVID-19
It has been found that immunological responses of circRNAs and lncRNAs may influence immunological tolerance and immune escape [75, 76]. In a study, authors found 898 differently expressed lncRNA in COVID-19-positive patients, of which 414 were up-regulated and 484 were down-regulated. Furthermore, among 570 circRNAs that had different expressions compared to healthy patients, 155 were up-regulated and 415 were down-regulated. Using circRNA/lncRNA as ceRNA may protect mRNA against microRNA depreciation [77]. LncRNAs affect cis-regulation in signal transduction such as Wnt/βcatenin, Ras, mTOR, and interleukin-1 mediated signaling pathways, which can affect transport, cell migration, phosphorylation of proteins, and protein transcription via repressing transcription factors. Furthermore, lncRNA expression in trans-regulation impacts drug metabolism. It also helps the host's natural immunity by affecting the assembly of the NLRP3 inflammasome complex [77].
7. LNCRNA AND EXOSOMES RELATIONSHIP
Studies also suggest that exosomes have a crucial function in viral infection [78]. Exosomes are small membrane vesicles that vary between 30 and 150 nm, carrying RNA and protein complexes in eukaryotic cells into the extracellular matrix [77]. Immune response, antigen presentation, cell proliferation, diversification, cancer development, and other processes may all be impacted by them, depending on the cell type from which they emerge. Exosomes can carry viral nucleic acid and proteins and can change the microenvironment, enabling diseases to propagate quickly [77]. Coronavirus-infected cells generate exosomes and may also export the SARS-CoV-2 invasion receptor (ACE2), rendering uninfected cells susceptible to virus loading [79]. CircRNA and lncRNA variants linked to exosomes were discovered in SARS-CoV-2 infected cells. Therefore, it has been proposed that exosomes might be used in COVID-19 treatment. A few studies have found that the composition of exosomes differs before and after infection, indicating that exosomes may be involved in developing new diseases. These exosomes contain 114 differently expressed circRNAs and 10 differentially expressed lncRNAs [80, 81].
8. LNCRNAS INTERACTION WITH IMMUNE SYSTEM COMPONENTS IN SARS-COV-2 INFECTION
The current SARS-CoV-2 pandemic's high morbidity and death required the development of efficient therapeutic techniques to fight the pathogenesis of SARS-CoV-2 [82, 83]. According to studies on SARS-CoV-infected humans and animals, the virus's fatal pneumonia may be linked to immune-pathological processes [84-86]. The significant COVID-19 pathology and rising clinical symptoms may be due to inflammation and cytokine storm [82, 83].
Multiple studies have shown that cytokines such as IL-1, IL-6, IL-12, IL-18, TNF, IFN, and other inflammatory mediators, are released uncontrollably in “Cytokine Release Syndrome” or CRS, which is believed to be linked to severe COVID-19 [87]. It is noteworthy that the function of these components of the immune system is through non-coding RNAs such as lncRNAs [32]. The INK4 gene's antisense non-coding RNA (ANRIL), which forms an endogenous competitive RNA, has been linked to inflammatory responses [88, 89].
8.1. IL-6 secretion-related lncRNAs
IL-6, a multifunctional cytokine that promotes acute inflammatory responses, influences many cancer types as well as viral infections [90, 91]. lncRNAs regulate IL-6 synthesis through several mechanisms involving JAK/STAT, NF-κB, HIF-1, and MAPK (Fig. 2) [92-95]. As an example, MALAT1 (metastasis-associated lung adenocarcinoma miRNA 1), also known as NEAT2, has been discovered to have a dual function in various signaling pathways, especially IL6 [96].
The Janus kinase/signal transducers and activators of transcription (JAK-STATA) are downstream signaling pathways that are triggered by IL-6 (STAT1,3, and 5), which affect immunological processes [97, 98]. Nuclear factor kappa-B (NF-κB) is important for IL-6 secretion and modulation of NF-κB, a major transcription factor of IL-6 that has been shown in preclinical trials to suppress SARS-CoV [99]. LncRNAs may influence IL-6 expression in a myriad of contexts, one of which is NF-κB [92-94]. Another long intergenic non-coding RNA (lincRNA) linked with inflammatory events through the NF-κB pathway is Gm4419 [100].
8.2. NLRP3 inflammasome Development by lncRNAs
The growing body of evidence indicates that inflammasome formation is aided by lncRNAs, which are linked to promoting severe diseases [101, 102]. The NLRP3 inflammasome is activated against infectious pathogens such as SARS-CoV-2, producing IL-1 and IL-18 [32, 62, 103]. Two different signaling pathways activate the NLRP3 inflammasome. First, pro-IL-1 and NLRP3 are enhanced by microbial compounds that recognize TLRs or cytokines and stimulate NF-κB. Pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) activate the secondary pathway, resulting in the construction of ASC and pro-caspase-1 and, as a result, the stimulation of the NLRP3 inflammasome [104, 105]. Therefore, it's becoming apparent that anti-NLRP3 inflammasome medications may benefit patients with inflammatory illnesses by reducing inflammatory responses [106-108].
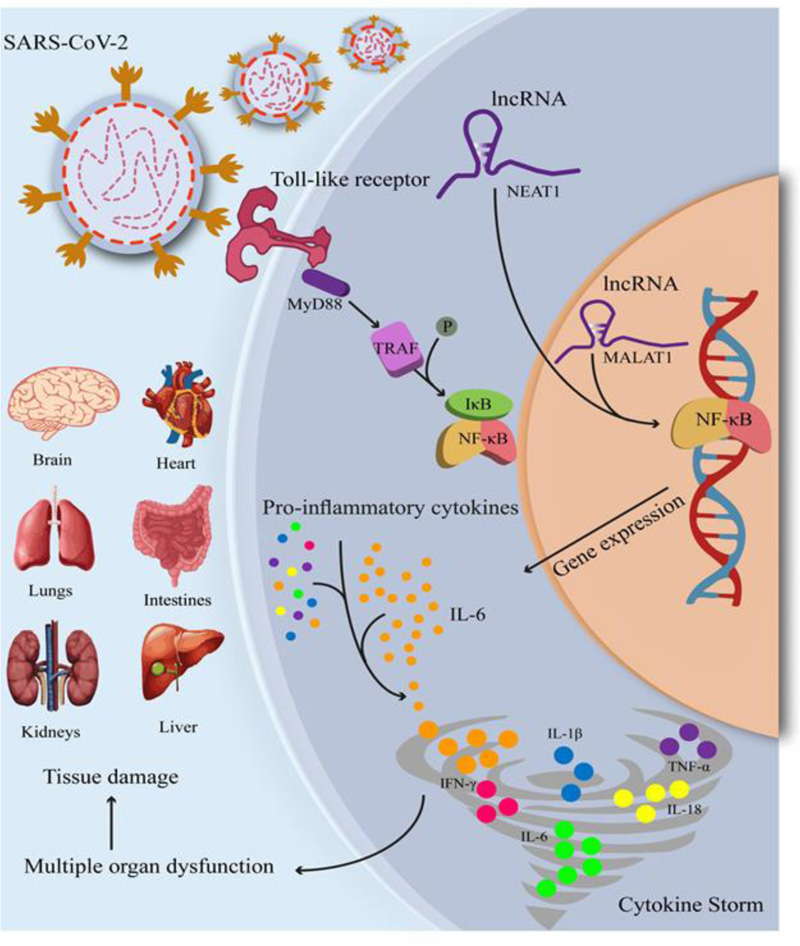
NEAT1 (nuclear enriched abundant transcript 1) has been associated with several different malignancies, including cancers such as prostate, cervical, and breast cancer [109]. Furthermore, LPS-stimulated immortalized bone marrow-derived macrophages (iBMDMs) enhanced NEAT1 translocation in the nucleus and cytoplasm, resulting in inflammasome assembly, caspase one activation, and inflammatory cytokine response, highlighting NEAT1's inflammatory role [45].
9. INFLAMMATION AND CYTOKINE STORMS: THE ROLE OF MICRORNAS
Ariana Centa et al. believe that microRNAs are involved in endothelial function in individuals suffering from serious respiratory problems and thrombotic complications in post-mortem lung tissues. The findings of a study indicate that miR (-26a-5p,-29b-3p, and-34a-5p) recognize mRNA targets implicated in endothelial and inflammatory signaling pathways as regulators, as well as viral diseases. Based on miRNA targets, protein-protein interactions, and inflammatory indicators found in the patients, the miRNA/mRNA network showed a strong relationship between these miRNAs and endothelial activation/dysfunction. MiR-26a-5p [IL-6 and ICAM-1] and miR-29b-3p [IL-4 and IL-8] have a significant relationship with inflammatory biomarkers in COVID-19 patients. The results showed miR (-26a-5p, -29b-3p, and -34a-5p) endothelial dysfunction and inflammatory response in people infected with SARS-CoV-2, as well as the development of severe lung damage and immunothrombosis [110].
The rapid and widespread edema and fibrosis that occur during the remodeling process and eventual airway clogs are undoubtedly caused by uncontrollable and abrupt increases in TGF-β [111]. MiR-27a-3p regulates TGF function, inhibits the TGF-β /Smad pathway, and suppresses myofibroblast development by regulating Smad2 and Smad4 activity [58].
Jacopo Sabbatinelli et al. predicted that COVID-19 severity is related to inflammation. They analyzed samples from COVID-19 individuals with multifocal interstitial pneumonia to examine their reaction to a single intravenous infusion of tocilizumab, an anti-IL-6 receptor drug (TCZ). They evaluated a set of microRNAs that control inflammation, miR-146a-5p, miR-21-5p, and miR-126-3p, with RT-PCR and Droplet Digital PCR techniques. As a result, COVID-19 patients who did not respond to TCZ exhibited lower blood levels of miR-146a-5p and they had the poorest outcomes. These findings suggest that a blood-based biomarker like miR-146a-5p may provide insight into the molecular link between inflammation and the clinical course of COVID-19, allowing us to understand better how to use biological drug armament to fight this global health issue [112].
In nanoparticle technology, using cerium oxide nanoparticles (CNP) allows unstable medicines, such as the anti-inflammatory microRNA-146a, to be administered locally to the damaged lung without causing systemic absorption. In a study, the intrathecal injection of CNP-miR146a showed an improvement in lung biomechanics by decreasing inflammation and oxidative stress via modulating leukocyte recruitment and reducing collagen deposition [113].
According to recent research, the IL-6/IL-6R pathway is a significant contributor to symptom-related cytokine storms. Downregulation of miR-451a may increase IL-6R protein production. In COVID-19 patients, three up-regulated long non-coding RNAs (lncRNAs) with miR-451a binding sites may act as miRNA sponges, competing with IL-6R for miR-451a. These results help researchers identify therapeutic targets for this novel illness [114].
Dharmendra Kumar Soni et al. investigated the pathogenicity of SARS-CoV-2 infection by examining the function of the most potent antiviral reactions in the host and immunological and inflammatory responses. The efficacy of anti-miR-155 treatment was tested in a COVID-19 animal model (mice transgenic for human angiotensin I-converting enzyme two receptors). Their findings show that male models had higher levels of viral loads and miR-155 than female ones. Furthermore, they found that treating SARS-CoV-2-infected mice with anti-miR-155 lowers miR-155 expression and improves survival and clinical outcomes. Anti-miR-155-treated mice hACE2 infected with SARS-CoV-2 exhibited improved antiviral and anti-inflammatory cytokine responses and reduced levels of pro-inflammatory cytokines [115].
In the latest research, differentially expressed circulating miRNAs have been recognized as viable biomarkers for understanding the severity of the disease in the Brazilian population using high-throughput sequencing to detect miRNA expression levels. A total of 18 human miRNAs were expressed differently in COVID-19 patients, with 13 miRNAs being substantially elevated and 5 miRNAs being considerably downregulated. Moreover, miR (-4433b-5p, -6780b-3p, -6883- 3p, -320b, -7111-3p, -4755-3p,-320c, and miR-6511a-3p) were shown to be significantly involved in the PI3K/AKT, Wnt/-catenin, and STAT3 signaling pathways, all of which are essential in viral infections. MiR-451a, -101-3p, -185-5p, -30d-5p, -25-3p, -342-3p, -30e-5p, -150-5p, 15b-5p, and 29c-3p were the most significant miRNAs found to be engaged in the Wnt/-catenin, NF- κB, and STAT3 signaling pathways, which play critical roles in immune response and inflammation. However, further studies are required to confirm these miRNAs as COVID-19 biomarkers [116].
9.1. MicroRNAs as Biomarkers for the Acute and Post-acute Phases of COVID-19
The outcomes of a real-time PCR assay were used to assess the expression level of selected miRNAs such as let-7b-3p, miR-29a-3p, -146a-3p, and 155-5p in peripheral blood mononuclear cells (PBMCs) of COVID-19 patients, in both acute and post-acute phases, and healthy controls. In COVID-19 patients, receiver operating characteristic analysis [86] was used to assess the specificity and sensitivity of miRNAs. All miRNAs were expressed at greater levels in COVID-19 patients. As a result, the expression patterns of miR-29a-3p, miR-146a-3p, and let-7b-3p were substantially different in the post-acute COVID-19 phase compared to the acute COVID-19 phase. ROC analysis recognized MiR-29a-3p, -155-5p, and -146a-3p as new biomarkers for COVID-19 diagnosis with excellent specificity and sensitivity. Furthermore, miR-29a-3p and -146a-3p may be used as novel biomarkers to differentiate between the acute and post-acute phases of COVID-19 [117].
9.2. MicroRNAs can Regulate the Expression of ACE2 and TMPRSS2 Receptor Genes
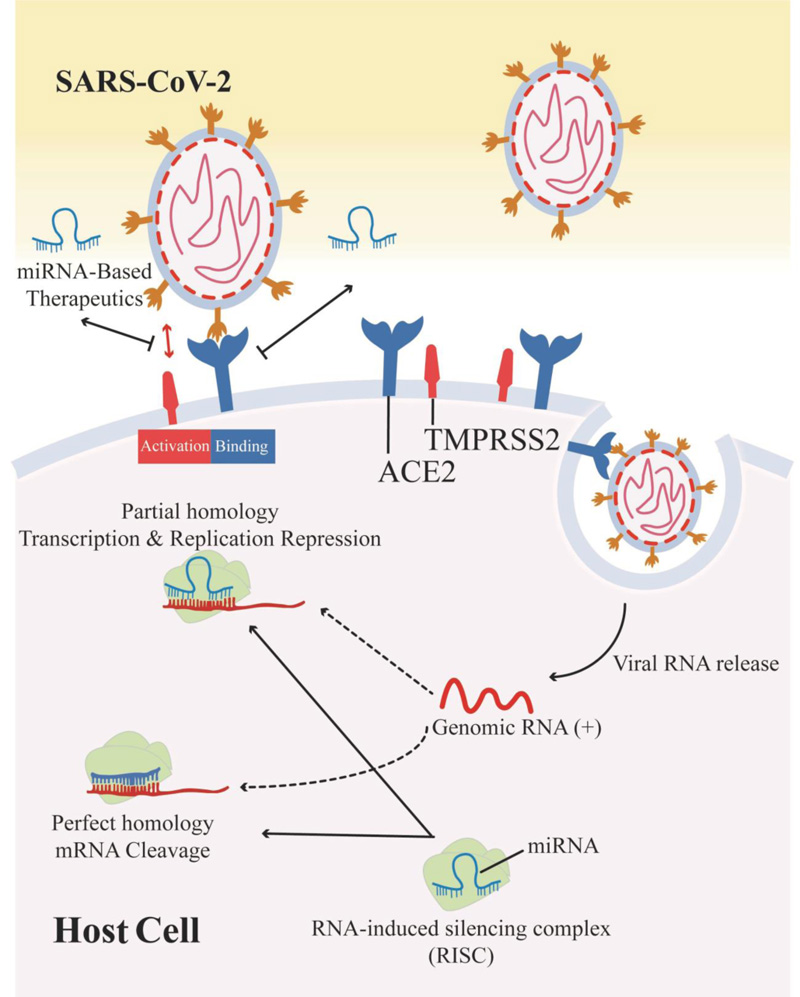
Zhi Liu et al. investigate the role of miRNAs in virus-induced dysregulation. According to their results, infection-modulated miRNAs regulate two of the most important biological processes: the immune response and cytoskeleton structure [118]. The control of cellular components, molecular activities, and biological processes was used to group all of the differentially expressed miRNA target genes, as determined via cluster analysis. According to enrichment analyses, peptidase, protein kinases, and the ubiquitin system exhibited the highest enrichment values [119]. SARS-CoV-2 enters cells by latching on to the receptor ACE2 with the spike (S) protein via the host serine protease TMPRSS2, which allows viral and cellular membranes to merge (Fig. 3) [55].
Given the significance of cellular receptors, particularly ACE2, in SARS-CoV-2 infection, Sardar et al. discovered that miRNA 27b controls the ACE2 receptor [120]. According to the findings of Chauhan et al., miRNA 200b-3p, miRNA 200c-3p, and miRNA 429 can inhibit ACE2, whereas let-7c-5p, miRNA 98-5p, let-7 f-5p, let-7 a-5p, let-7 g-5p, let-7 g-5p, let-7b-5p, miRNA 4458, let-7e-5p, let-7i-5p, let-7d-5p, and miRNA 4 can increase the expression of ACE2. Increased expression of the ACE2 receptor is observed in patients with metabolic syndrome, diabetes, and heart disease. Therefore, inhibiting the ACE2 receptor with miRNAs may be an effective treatment option for COVID-19 infection [121].
In addition to miRNAs that directly interact with the viral genome, host miRNAs that target ACE2 may play a role in regulating SARS-CoV-2 infection. In addition, a large number of miRNAs targeting the 3′-UTR of ACE2 in humans were discovered. All three online miRNA prediction algorithms found six miRNAs (miR-362-5p, miR-421, 500a-5p, 500b-5p, miR-3909, and 4766-5p). MiR-421 has already been identified as a possible ACE2 regulator, which is interesting [122].
There has been evidence of miRNA dysregulation in patients with SARS-CoV-2, which may cause changes in the genes controlled by miRNAs [41]. High-throughput sequencing was used to assess the expression levels of different miRNAs, and correlation analysis was used to discover the miRNA-primed target genes. Compared to healthy controls, 35 miRNAs were up-regulated and 38 miRNAs were downregulated in COVID-19 patients. The production of miR (6501-5p and 618) was 1.5-fold higher in COVID-19 patients as compared with healthy control donors. A 2.3-fold reduction in miR-627-5p, on the other hand, was seen in comparison to the controls. There was a 1.3-fold reduction in the expression of other miRNAs (miR-183-5p, 627-5p, and 1443-3p) compared to healthy donors. They found that hsa-miR-4661-3p, a virus-encoded miRNA, SARS-S CoV-2's gene was anticipated to be the target [118].
According to a study, SARS-CoV-2 needed miR-200c to connect ACE2 receptors in cardiomyocytes [123]. To identify common regions and coronavirus 2 genes linked to SARS (Severe Acute Respiratory Syndrome), Abolfazl Bahrami and Maryam Bakherad retrieved the entire genomes of all viruses identified in databases for this family (55 genomes in all) and conducted comparative genomic research on them. RELA in the viral genome and ACE2 receptors and CLEC4M genes in the host genome were the most significant genes implicated in the illness. RELA gene was reduced by hsa-miR (516b-3p, 3529-3p, and 6749-3p), ACE2 receptor was inhibited by hsa-miR (23b-5p and 769-5p), and CLEC4M gene was repressed by hsa-miR (4462 and 5187-5p). As a consequence, their findings will aid in the management and treatment of COVID-19, as well as provide fresh insight into vaccine design and miRNA therapy [124].
The NF-κB pathway activation raises the expression of miR-200c-3p, which is a key factor in ARDS. The increased miR-200c-3p expression has been linked to a reduction in ACE2 expression. In certain COVID-19 instances, reduced ACE2 expression in the lungs and upper respiratory tract may be related to decreased disease severity. As a result, it is postulated that bacterial LPS and LTA may lower ACE2 expression in COVID-19 patients' lungs via up-regulating miR-200c-3p [125, 126].
Transmembrane serine protease 2 (TMPRSS2) has an important role in mediating viruses, and SARS-CoV-2 employs TMPRSS2 for viral entry [127]. SARS-CoV-2 can encode miRNAs that promote TMPRSS2 overexpression, and Mir-147-3p can increase SARS-CoV-2 infection in the gut by boosting the synthesis of TMPRSS2 [118]. However, miRNAs may be used as molecular tools to prevent SARS-CoV-2 viral transmission and replication in humans since they have a strong affinity for TMPRSS2 and may block this receptor [128]. TMPRSS2 diminution mediated by miRNA for early COVID-19 prevention has been established in the laboratory for early COVID-19 prophylaxis. TMPRSS2 binding was examined in a pool of 163 miRNAs using three miRNA prediction methods, yielding 11 common miRNAs. Furthermore, negative computational energies for association confirmed miRNA-Tmprss2 interactions, while the S fold tool identified three miRNAs (hsa-miR-214, hsa-miR-98, and hsa-miR-32) based on likelihood scores of 0.8 and accessibility to the Tmprss2 target. Transfection of miRNA(s) into Caco-2 cells, quantitative differential expression analysis, and confirmation of Tmprss2 silencing with maximal gene suppression by hsa-miR-32, is a new potential function in CoV-2 pathogenesis [128].
CONCLUSION
Many of the strategic uses of non-coding RNAs to promote SARS-CoV-2 may be unrecognized. As a result of viral infections, the regulation of lncRNAs occurs irregularly, and many host functions are improperly regulated, leading to the development of viral infection. Human non-coding RNAs are influenced by viral infection as viral proteins interact intimately with their host proteome. On the other hand, A low-scale approach to studying non-coding RNAs in response to SARS-CoV-2 infection could be inappropriate in the present circumstances. We introduced many lncRNAs with altered expression levels during viral infection, which may serve as biomarkers. However, their specific role in response to the virus is yet to understand.
Furthermore, microRNAs play significant roles in COVID-19, and the presented strategies might lead to the creation of procedures for reducing the expression of critical COVID-19 “cytokine storm” components. The characteristics of lncRNA and miRNA can be employed in the treatment and diagnosis of patients with the SARS-CoV-2 virus. However, additional studies are required.
LIST OF ABBREVIATIONS
| ncRNAs | = non-coding RNAs |
| COVID-19 | = Coronavirus Disease 2019 |
| ceRNAs | = Competing for Endogenous RNAs |
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
None declared.


